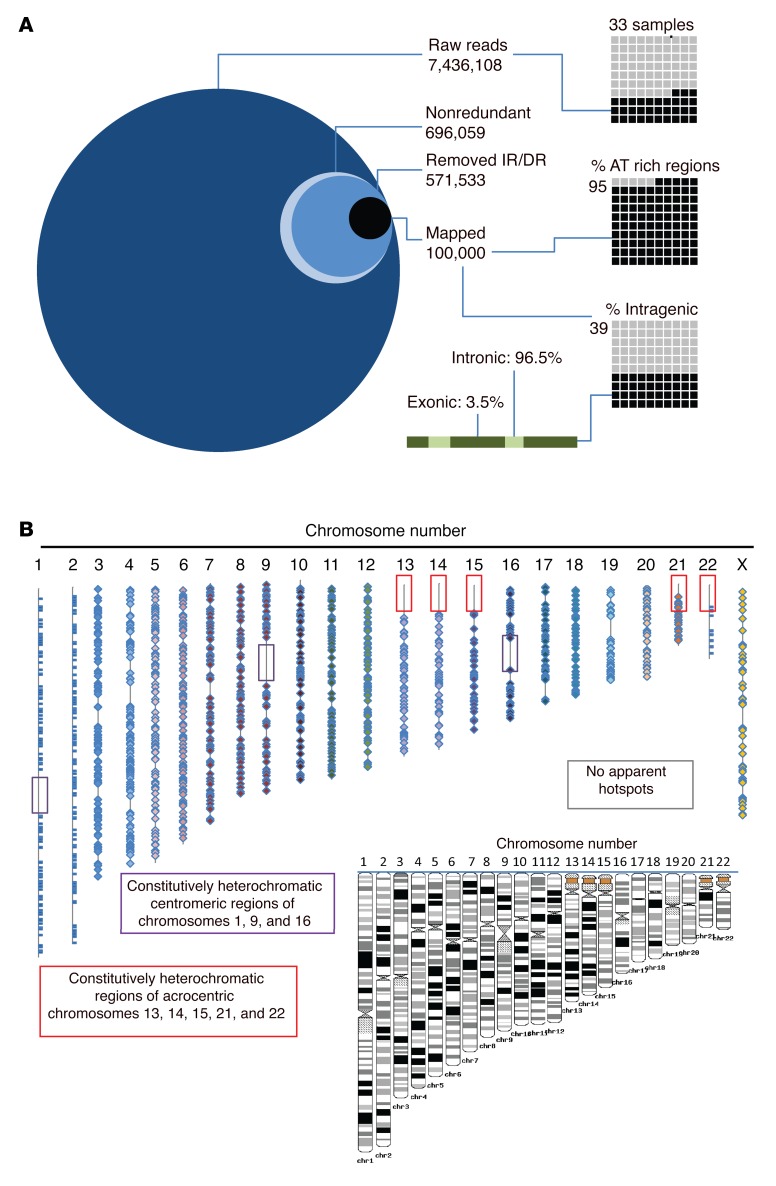
Figure 1. High-throughput sequencing reveals distribution and genomic location of CAR integrants after SB-mediated transposition in primary T cells.
(A) The integration site for CAR insertions was determined in genomic DNA libraries (n = 33) isolated from independent T cell populations following genetic modification with the SB system and propagation on AaPCs with cytokines to stably express CD19RCD28 CAR. From greater than 7 million raw sequences, 696,059 nonredundant sequences were obtained. The inverted repeats (IRs) and direct repeats (DRs) were digitally removed, leaving 571,533 unique integrations, of which 100,000 were mapped. The percentages in AT-rich regions and in intragenic regions are displayed in the right-hand graphics. The green bar and associated text display the percentage of intragenic integrations located in introns (light green) vs. exons (dark green). (B) The location of each SB integration (CAR) is mapped onto the 23 human chromosome scaled (as shown in black and white inset) representations. Boxes denote constitutive heterochromatic regions that could not be analyzed. Each integration is noted with a bar (chromosomes 1, 2, and 23) or diamond. Integrations are widely dispersed throughout the genome, without hotspots.