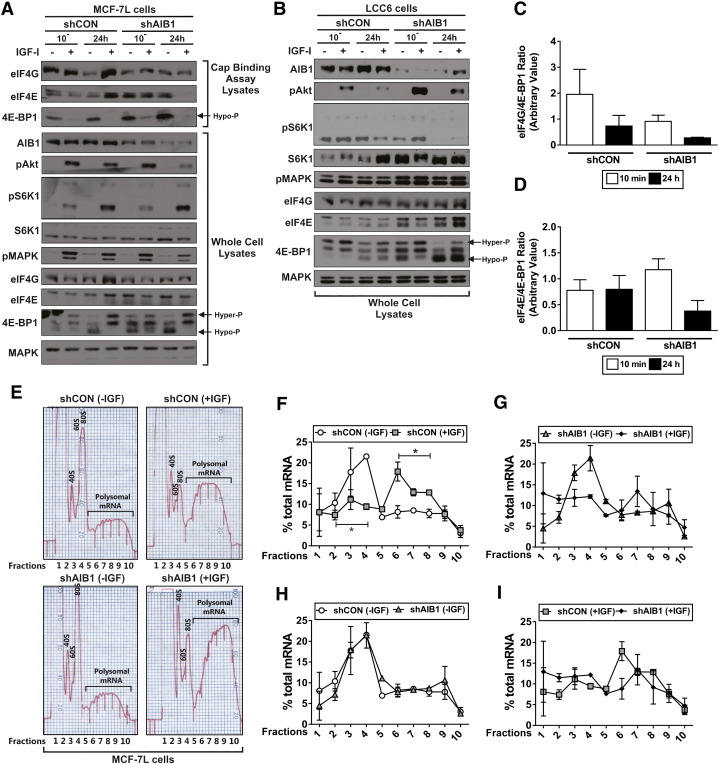
Figure 2.
AIB1 knockdown increases eIF4E and 4E-BP1 complex steady-state protein levels and reduces polyribosome recruitment.
A) Micro-scale affinity chromatography using m7GTP agarose beads (top) was performed using shRNA scrambled control (shCON) or AIB1 (shAIB1) IGF-I treated MCF-7L cell protein lystates. Immunoblot analysis was performed on whole cell protein lysates (bottom). Hyper-phosphorylated (hyper-P; top bands) and hypo-phosphorylated (hypo-P; lower bands) of 4E-BP1 are depicted on the immunoblot images in A and B.
B) Immunoblot analysis was performed on whole cell protein lysates obtained from shCON and shAIB1 LCC6 IGF-I treated cells.
C and D) Densitometry was performed on the m7 GTP cap-binding assay MCF-7L immunoblots to determine a relative translational index by calculating the eIF4G/4E-BP1 and eIF4E/4E-BP1 ratios of control cells for 10 minutes and 24 hours.
E) Densitometry (O.D.254) traces of polysome preparations. 107 shCON and shAIB1 MCF-7L cells were serum starved in PRF-media (plus additives) for 24 hours and subsequently treated +/− 5 nM IGF-I for 24 hours. Total RNA was isolated, loaded onto sucrose gradients, ultracentrifuged to stratify the ribosome bound mRNA transcripts based on the number of bound ribosomes, and fractionated into 10 fractions. Fraction 10 represents transcripts with the most bound ribosomes. Free ribosomal subunits (40S and 60S), monosomes (80S) and the polysomal mRNA fractions are depicted on the densitometry trace readings.
F-I) Percent mRNA per individual fraction normalized to total polysome bound RNA. Experiments were performed three times with similar results; graphs represent combined data from two experiments. Error bars are SEM *P < .05 one-way ANOVA. A weighted average calculation was performed to demonstrate an overall shift in the average number of bound ribosomes.