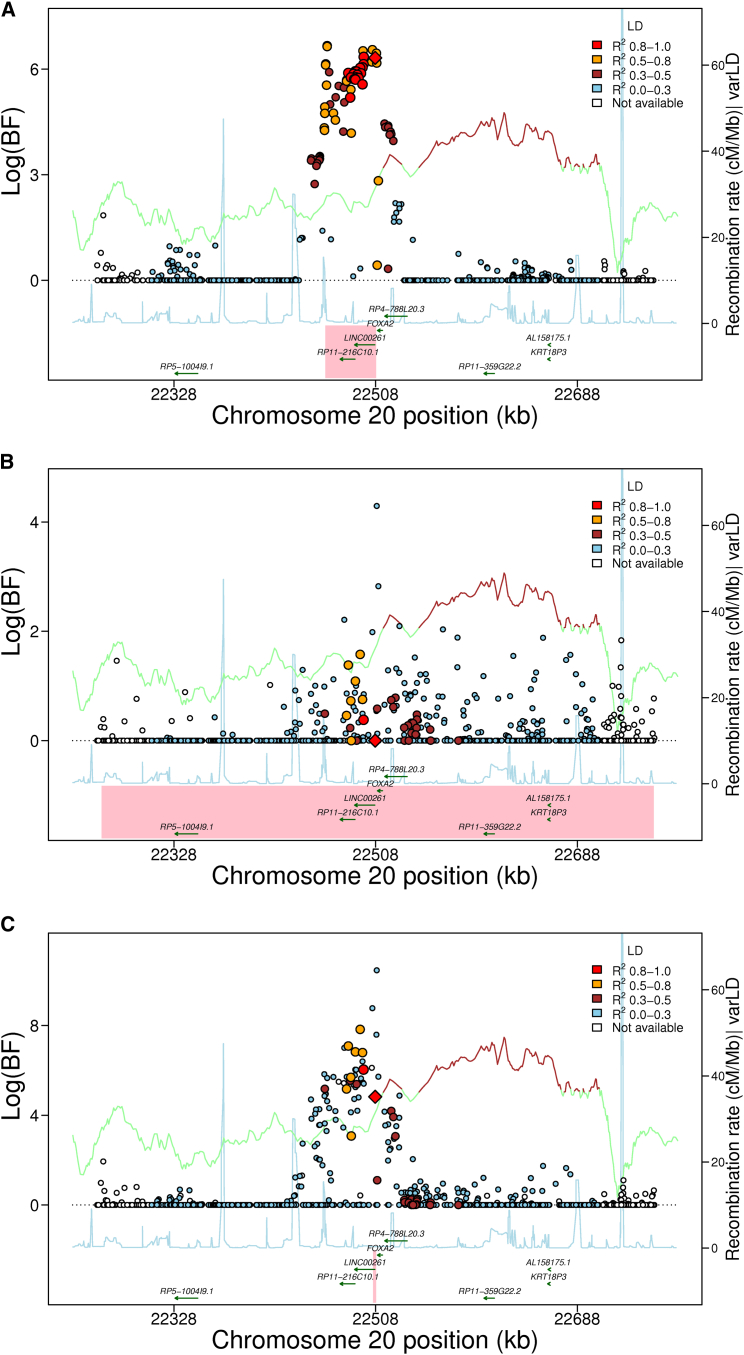
Figure 1.
Trans-ethnic Analysis of Glycemic Quantitative Loci Provides Narrowed Intervals Spanned by the 99% Credible Set
Data are 500 kb regional association plots for fasting glucose at FOXA2, centered at the index SNP identified from European ancestry (EA) samples. The x axis denotes genomic position and the y axis denotes the log (BF), recombination rate, and varLD information61 (a measure to quantify LD variation differences comparing populations). The red diamond data point represents the index SNP within the region previously reported from EA samples. The color of each data point indicates its LD value (r2) with the index SNP based on HapMap2 (YRI for AA results and CEU for EA results): white, r2 not available; blue, r2 = 0.0–0.2; brown, r2 = 0.2–0.5; orange, r2 = 0.5–0.8; red, r2 = 0.8–1.0. The blue line represents the recombination rate. The green line shows the varLD score at each SNP and is highlighted with dark brown if the varLD score is >95th percentile of the genome-wide varLD score, comparing LD information between YRI and CEU HapMap2 samples.61 The interval spanned by the 99% credible set is highlighted in pink.
(A) Association results for fasting glucose in the FOXA2 region in EA individuals. The 99% credible set contains 40 SNPs that span an interval of 46,365 bp.
(B) Association results for fasting glucose in the FOXA2 region in AA individuals. The association signal is weaker than in EA samples, leading to a wider interval spanned by the 99% credible set.
(C) Association results for fasting glucose in the FOXA2 region in both EA and AA individuals. The 99% credible set contains 2 SNPs and spans an interval of 3,872 bp, a 95% reduction in the number of SNPs and a 91.6% reduction in the length of the credible set interval.