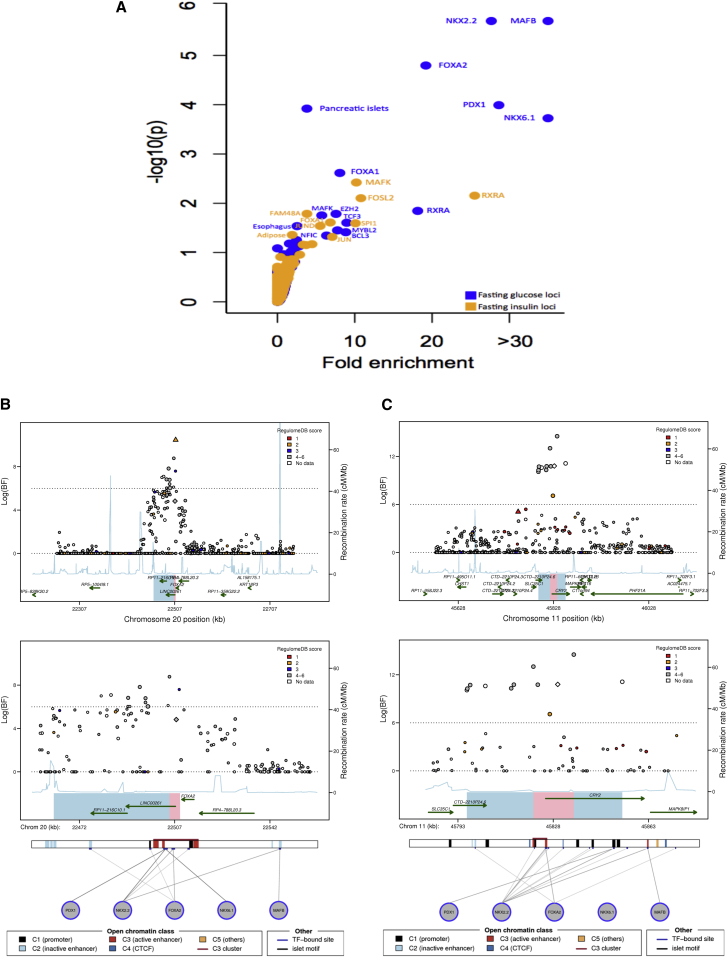
Figure 2.
Trans-ethnic Fine-Mapping of Glycemic Quantitative Trait Loci Highlights Overlap between Trait-Associated SNPs and Predicted Regulatory Function
(A) Analysis for enrichment of posterior probabilities in SNPs overlapping transcription factor binding sites (TFBSs) and cell-type-specific enhancer and promoter marks at 22 (13 FG, 8 FI, and 1 both FG and FI) substantially narrowed 99% credible sets. x axis shows fold-enrichment above null, the y axis shows –log10(P) for enrichment, and FI and FG are indicated by yellow and blue points, respectively. TFBSs and cell types with enhancer or promoter marks with p value for enrichment below 0.01 are labeled.
(B) Regional association plots for fasting glucose after trans-ethnic analysis demonstrating overlap between regulatory annotation and narrowed credible regions at the FOXA2 locus.
(C) Regional association plots for fasting glucose after trans-ethnic analysis demonstrating overlap between regulatory annotation and narrowed credible regions at the CRY2 locus.
(B and C) The index SNP in European ancestry (MAGIC) is represented by a diamond; the best SNP in African ancestry (AAGILE) is represented by a triangle. SNPs are colored according to the score assigned in RegulomeDB11 with lower score corresponding to stronger level of evidence supporting regulatory function; data from the Islet Regulome Browser13 for the genomic interval is shown below regional association plots. The interval spanned by the 99% credible set using EA data only is represented by combining blue and pink regions; interval spanned by the narrowed 99% credible set after trans-ethnic analysis is shown in pink.