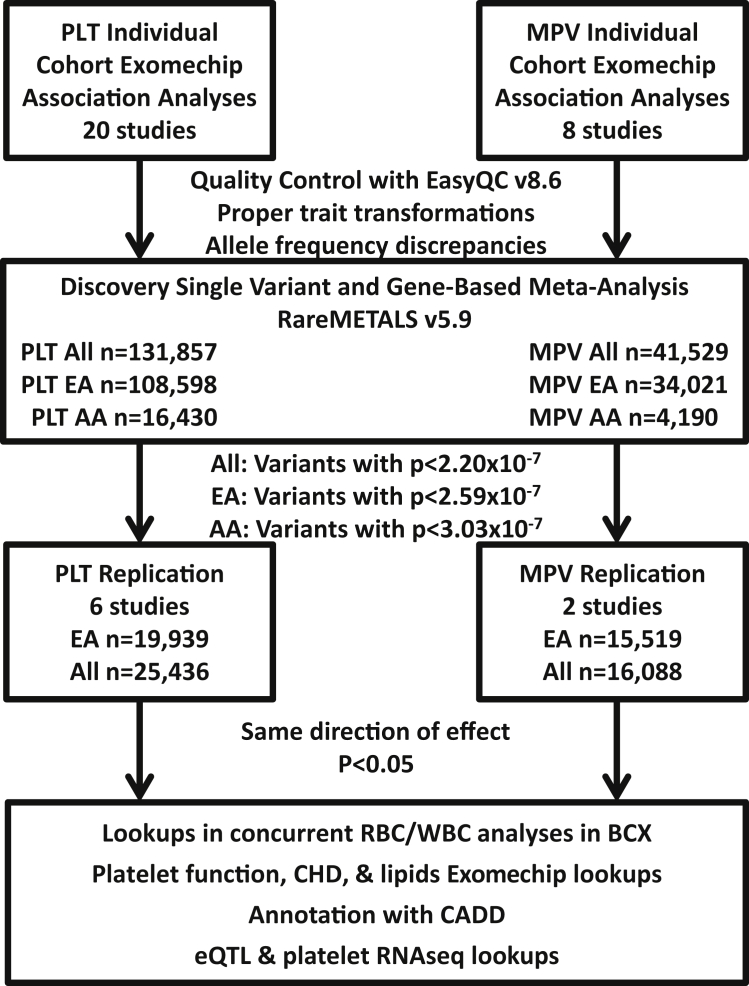
Figure 1.
Study Design and Flow
Individual study-level association analyses were performed using RareMetalWorker or RVTests. To perform quality control of individual study association results, we used EasyQC v.8.6 to ensure proper trait transformations, to assess allele frequency discrepancies, and to evaluate other metrics. We then combined results in meta-analysis with RareMETALS v5.9 in three groups: African ancestry (AA), European ancestry (EA), and combined all five (AA, EA, Hispanic-Latino, East Asian, South Asian) ancestries (All). Independent variants identified by conditional analysis in RareMETALS with a p value less than the threshold corrected for multiple testing (All, p < 2.20 × 10−7; EA, p < 2.59 × 10−7; AA, p < 3.03 × 10−7) were carried forward for replication. Markers showed replication if they had p < 0.05 in the same direction of effect in the replication analyses. Associated markers were further annotated using various resources: (1) concurrent BCX Exomechip analyses of RBC and WBC traits, (2) on-going Exomechip analyses of platelet aggregation, quantitative lipids, and coronary heart disease (CHD) traits, (3) severity prediction by CADD, (4) an internal database of reported eQTL results, and (5) platelet RNA-seq data.