Figure 3.
Mutations in CDC45 and Conservation of Substituted Residues in Orthologous Proteins
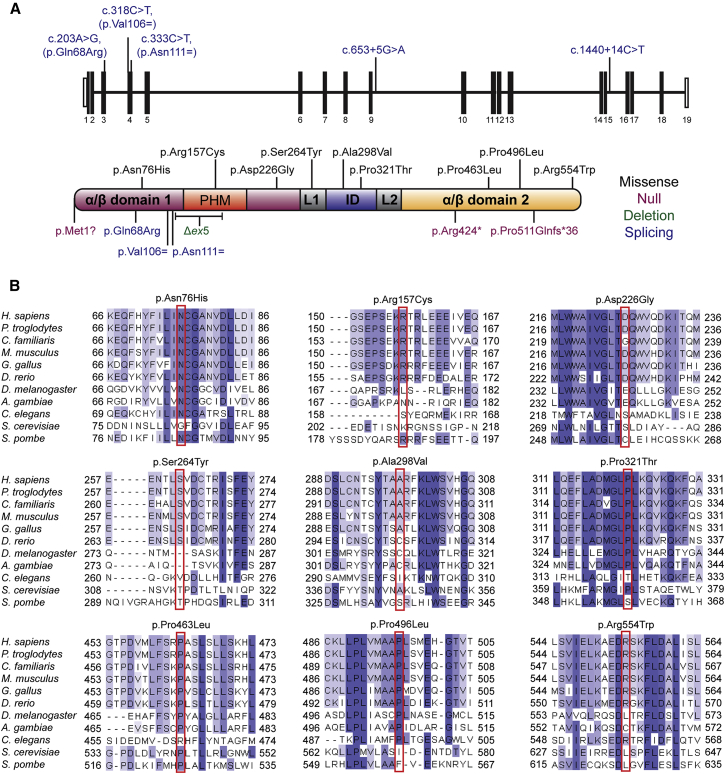
(A) Mutations identified in CDC45 include variants predicted or shown to affect splicing (blue, position in gene shown in upper panel), predicted missense substitutions (black), or truncating mutations (purple). Δex5 refers to the intragenic deletion encompassing exon 5 of CDC45 in family P12 (green). Mutations do not cluster within any specific domain of CDC45. Domain architecture of human CDC45 indicated as described in Yuan et al.26 The two alpha-beta domains are shown in purple and orange, respectively. The protruding helical motif (PHM) inserted in the first alpha-beta domain is shown in red. The middle all-helix interdomain (ID) is shown in blue, and the two linkers (L1 and L2) are shown in gray.
(B) Clustal Omega alignments of eukaryotic CDC45 proteins indicate mutated residues are well conserved, with several residues (such as Asn76, Arg157, Pro321, and Pro463) conserved as distantly as fungi. Blue shading indicates proportion of conserved amino acids.