Fig. 4.
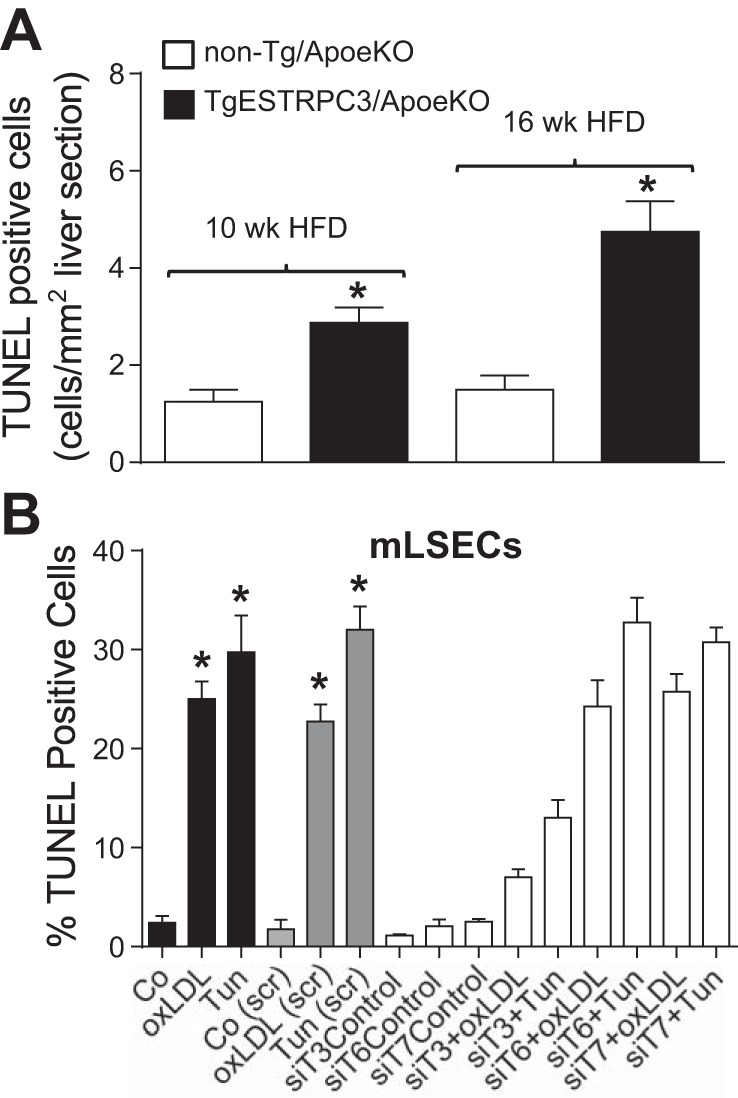
In A, liver sections (10 μm) from TgESTRPC3/ApoeKO mice or nontransgenic littermates (non-Tg/ApoeKO) that were maintained on a high-fat diet for 10 or 16 wk were stained for in situ terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL), and the number of apoptotic (TUNEL+) cells were counted and normalized per mm2 of section area. Shown are means ± SE (n = 5), *P < 0.05 vs. corresponding control. In B, liver sinusoid endothelial cells isolated from ApoeKO mice (mLSECs) were incubated in EBM-2 (Control) or EBM-2 containing oxidized LDL (oxLDL, 50 μg/ml) or tunicamycin (Tun, 10 μg/ml) for 24 h and processed for TUNEL assay (black bars). Alternatively, mLSECs were transfected with scrambled oligonucleotides (gray bars) or with siRNA specific for TRPC3, 6, or 7 (siT3, siT6, or siT7; empty bars) and 48 h posttransfection subjected to the above described treatments and processed for TUNEL assay. Shown are means ± SE (n = 3). *P < 0.001 vs. corresponding control. Differences for “siT3+oxLDL” and “siT3+Tun” vs. corresponding scrambled-transfected controls have P < 0.0001. In the presence of siT6 or siT7, the effect of oxLDL and Tun was not different from corresponding scrambled-transfected controls.
