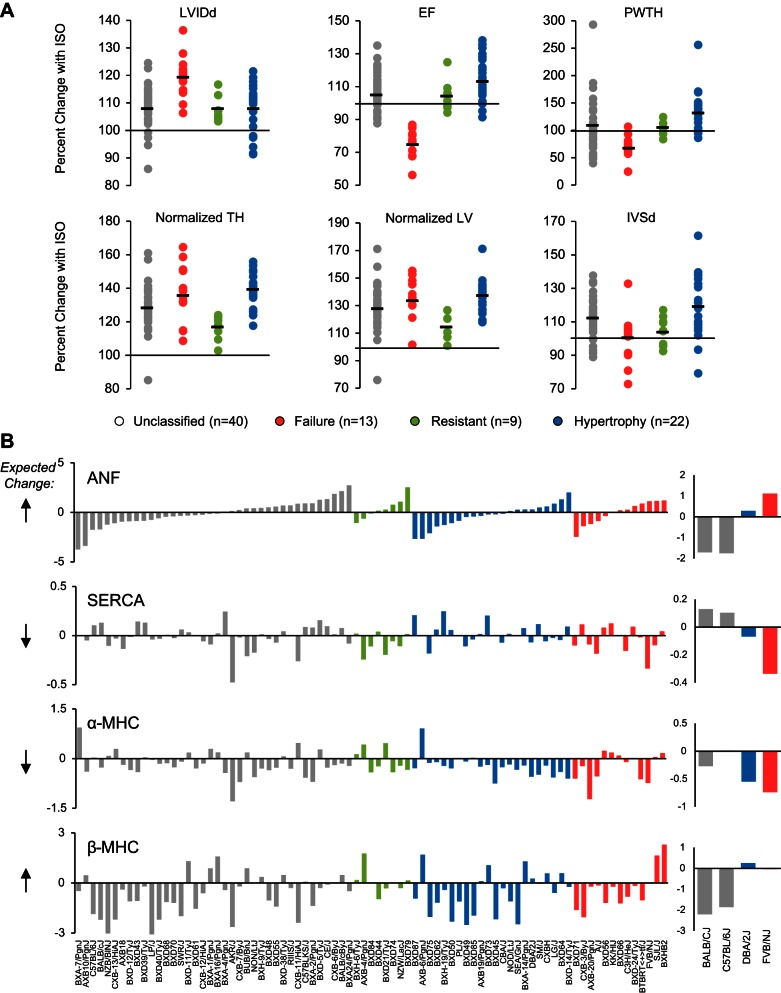
Fig. 1.
The fetal gene program does not coincide with cardiac hypertrophy in diverse genetic backgrounds. A: 84 inbred strains of mice were phenotyped after treatment with the beta-adrenergic agonist isoproterenol. Mice were categorized as hypertrophic or failing if the majority of their traits matched human clinical measurements of these conditions. Mice with minimal change after isoproterenol were classified as resistant. Strains whose traits were congruent with different conditions were left unclassified. Each dot represents a strain. Bar represents group mean. LVIDd, left ventricular internal diameter in diastole; EF, ejection fraction; PWTH, posterior wall thickness; normalized TH, total heart weight/body weight; normalized LV, left ventricular weight/body weight; IVSd, interventricular septum thickness at diastole. B: cardiac microarray data from the same 84 strains of mice were used to analyze change in expression of the fetal genes. With the exception of alpha myosin heavy chain (α-MHC), there was an equal number of strains upregulating or downregulating the fetal genes, even when controlling for disease state, suggesting they are not good markers of isoproterenol-induced hypertrophy across genetic backgrounds. Each bar represents a strain. Expected direction of change in expression is represented by the black arrows to the left. The commonly studied mouse strains BALB/cJ and C57BL/6J do not display expected trends in expression, while DBA/2J and FVB/NJ do (right panels). ANF, atrial natriuretic factor; SERCA, sarcoplasmic reticulum calcium ATPase.