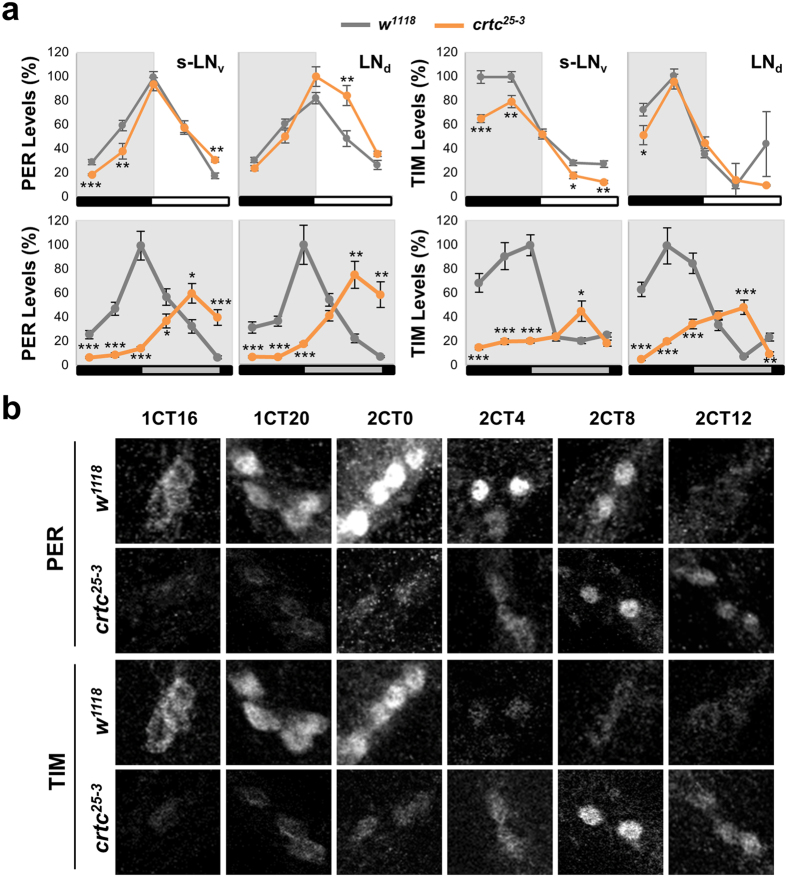
Figure 4. crtc mutation impacts on PER and TIM oscillations in circadian pacemaker neurons.
(a) Circadian expression of PER and TIM proteins in circadian pacemaker neurons of wild-type (gray lines) and crtc mutants (orange lines). Adult fly brains were dissected at different time-points in LD cycles (top) or during the transition from the first to the second DD cycle following LD entrainment (bottom). Whole-mount immunostaining was performed using anti-PER, anti-TIM, and anti-PDF antibodies. Confocal brain images were obtained from 12–13 hemispheres at each time-point (X-axis). The fluorescence intensity of anti-PER and anti-TIM antibody staining was quantified from individual neurons using ImageJ software and averaged for each group of circadian pacemaker neurons. Y-axis indicates the relative expression levels (%) of PER and TIM proteins, calculated by normalizing to the peak value in each graph (set as 100). White/black bars, LD cycles; gray/black bars, DD cycles. s-LNv, PDF-expressing small ventral lateral neurons; LNd, dorsal LN. *P < 0.05, **P < 0.01 and ***P < 0.001 as determined by Student’s t-test. Error bars indicate SEM. (b) Representative confocal images of small LNv in wild-type (w1118) and crtc mutants (crtc25-3). 1CT/2CT, circadian time in the first/second DD cycle.