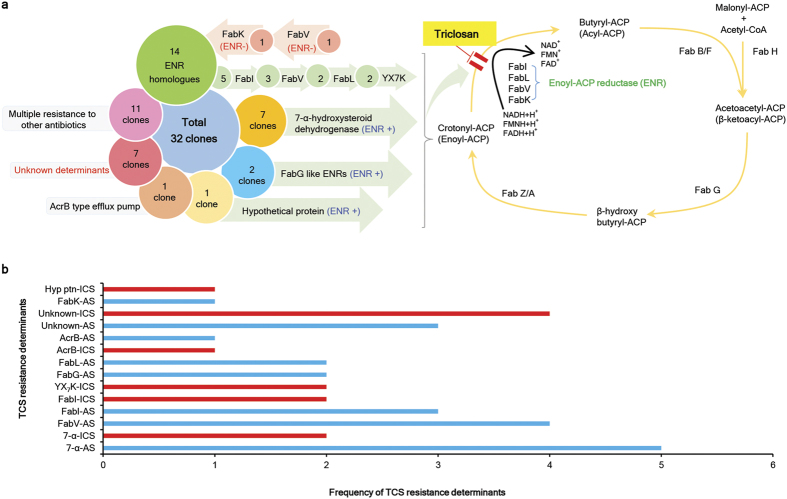
Figure 1. Metagenomic analysis of TCS-resistant clones revealed diverse patterns of TCS resistance determinants, with a higher diversity observed in AS compared to ICS.
(a) A summarized sketch of the TCS resistance determinants from metagenomic library screening. The majority of the clones carried different versions of ENR homologues, including previously known prototypic ENRs, novel candidate ENRs, and ENR homologues with compromised ENR activity. The ENRs that complemented ENR activity in vivo are indicated by forward light green arrows or are indicated otherwise. Other clones carried TCS resistance efflux pumps, hypothetical proteins or unknown determinants for resistance to this biocide. Some of the clones even carried genes for resistance to other antibiotics colocalized with the TCS resistance determinants. (b) Distribution of TCS resistance determinants between AS and ICS. TCS resistance determinants in AS and ICS are indicated in cyan and red, respectively. The comparative diversity of TCS resistance determinants was higher in AS compared to ICS.