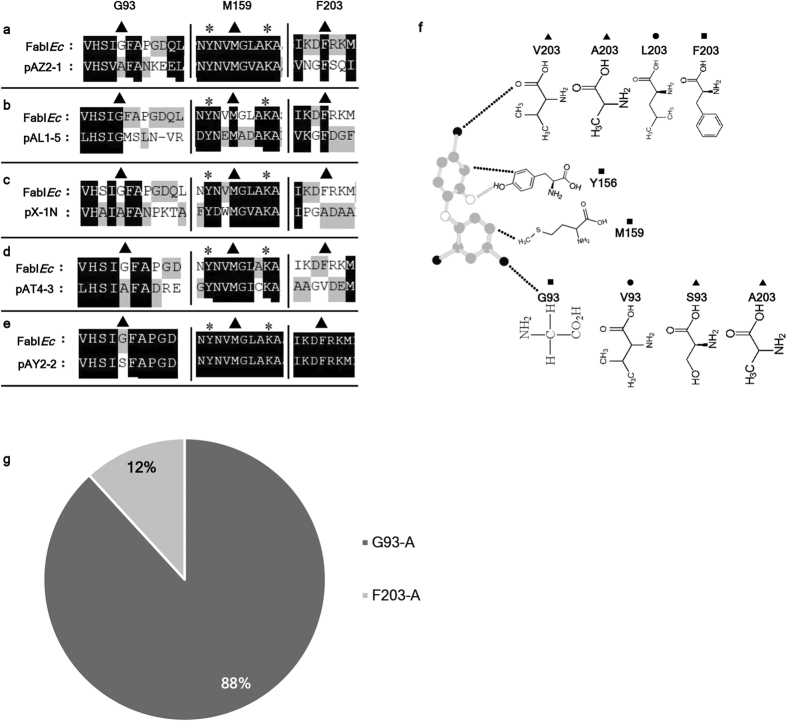
Figure 3. Metagenomic TCS-resistant FabI ENR homologues revealed unique patterns of substitutions that were abundant in most of the human-associated pathogenic and nonpathogenic bacterial FabI ENR homologues.
(a–e) Multiple alignment of metagenomic FabI ENR homologues with E. coli FabI ENR (on top). Only the multiple alignment regions carrying substitutuions associated with TCS resistance are presented here. Additionally, each original alignment has been divided into three sections, each separated by a vertical line, with individual alignments for each different ENR separated by horizontal lines. The YX6K-type active sites are marked with asterisks. Important residues whose mutations are associated with TCS resistance are indicated with closed triangles. (f) Schematic drawings of TCS interactions with important residues of the FabI ENR that are associated with TCS resistance. Ball-and-stick representation of TCS, with gray, black, and open white balls indicating carbon, chloride, and oxygen, respectively. Hydrogen bonds between TCS and amino acids are indicated by open white dotted lines, and hydrophobic interactions are indicated by black dotted lines. The neutral and mutated residues in the case of prototypic FabI are indicated with black rectangles and black circles, respectively. Amino acid substitutions found in the metagenomic ENRs are indicated with black triangles. Metagenomic TCS resistance associated amino acid substitutions carried different side chains. The variation in amino acid side chains at these positions are previously reported to be associated with triclosan tolerance16. (g) Metagenome-derived TCS-resistance-associated substitutions were dominant in FabI homologues of 401 pathogenic and non-pathogenic bacterial strains (122 bacterial species) (Supplementary Data 1). The G93-A substitution was more commonly present in 50% of bacterial ENRs, followed by the F203-A substitution, which was present in 7% of the bacterial ENRs from the investigated 401 bacterial strains. Therefore, the G93-A and F203-A substitutions constitute 88% and 12% of those found in ENRs, respectively.