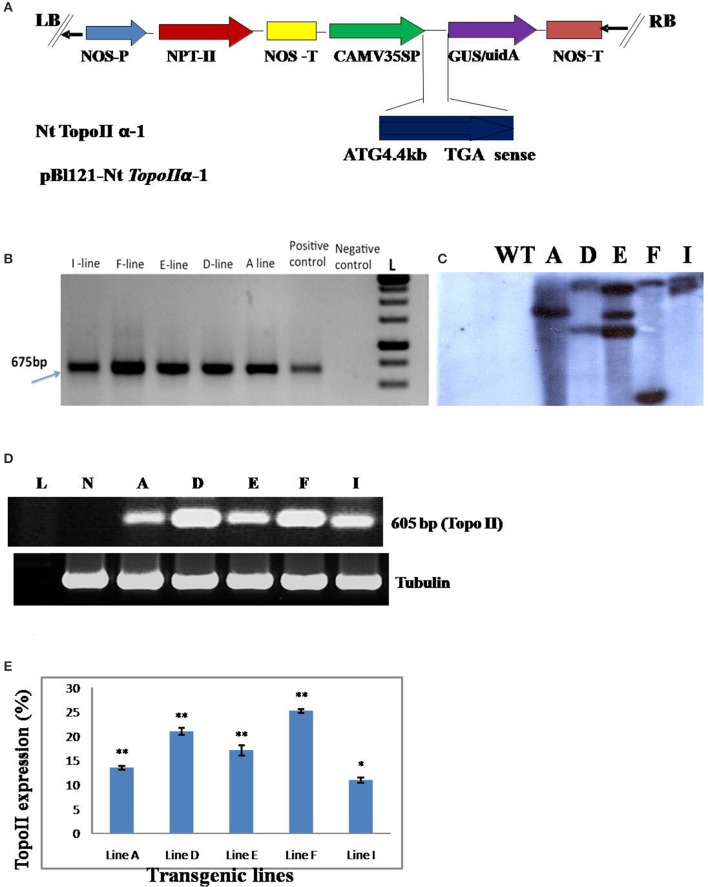
Figure 2.
(A) T-DNA map of NtTopoIIα-1 construct used for tobacco transformation. NOS P, nopaline synthase promoter; NPT-II, neomycin phosphotransferase II; NOS T, nopaline synthase terminator; CaMV P, Cauliflower mosaic virus 35S promoter; GUS uidA, β-glucaronidase; (B) Genomic DNA PCR analysis of untransformed control and putative TopoII transgenic tobacco plants (A–I lines) using npt-II gene-specific primers. L, DNA Ladder; Positive control, Plasmid DNA; Negative control, DNA from untransformed tobacco plant; (C) Southern blot analysis of transgenic and wild-type (WT) tobacco plants determining the T-DNA copy number. Genomic DNA was digested with XhoI, separated by electrophoresis on a 1% agarose gel and transferred onto a nylon membrane. The transferred DNA was hybridized with the npt-II gene probe labeled with α-32P dCTP. UT, DNA from the untransformed tobacco plant, A–I, transgenic lines; (D) Transgene expression (transcript) levels quantified by RT-PCR. Tubulin was used as an internal control. L, DNA ladder; N, Blank; A–I, transgenic lines; (E) The histograms represent the average of gene expression measured with the ImageJ, densitometry software. Values are the means of 3 replicates with the corresponding standard error. Bars labeled with asterisks show significant differences from that of WT at *P < 0.05 or **P < 0.01 by t-test.