Abstract
Background
Colorectal cancer (CRC) is the fourth leading cause of cancer-related death around the world and accumulated evidence indicates the association between CRC and obesity and insulin resistance.
Objectives
Regarding the role of adiponectin in obesity and insulin resistance, we explored whether genetic variants in adiponectin (ADIPOQ) and adiponectin receptor 1 (ADIPOR1) are associated with CRC risk.
Materials and methods
ADIPOQ (rs2241766) and ADIPOR1 (rs2275738) gene variants were genotyped in 261 cases with CRC and 339 controls using PCR-RFLP method.
Results
In this study, no significant difference was observed for ADIPOQ gene rs2241766 variant between the cases and controls. However, carriers of the ADIPOR1 (rs2275738) “CC + CT” genotype compared with “TT” genotype occurred more frequently in the cases with CRC than the controls, and the difference remained significant after adjustment for age, BMI, sex, smoking status, NSAID use, and family history of CRC (P = 0.048; OR = 1.49, 95%CI = 1.01–2.20). Interestingly, after adjustment for confounding factors the ADIPOR1 “CC + TC” genotype compared with “TT” genotype was also associated with an increased risk for obesity in the cases (P = 0.040; OR = 1.86, 95%CI = 1.03–3.36).
Conclusions
Our findings suggest for the first time that the − 106 C > T (rs2275738) variant of ADIPOR1 gene may be a genetic contributor to CRC and obesity risk in the cases with CRC. However, further studies with bigger sample size are needed to validate these findings.
Keywords: ADIPOQ, ADIPOR1, Colorectal cancer, Gene polymorphism
1. Introduction
Colorectal cancer (CRC) is the second most commonly diagnosed cancer and the fourth leading cause of cancer-related mortality across the world (Ferlay et al., 2010). Epidemiological evidence has shown that CRC is associated with obesity (Caan et al., 1998, Schoen et al., 1999), hyperinsulinemia and insulin resistance (Schoen et al., 1999, Trevisan et al., 2001). Previous studies have also demonstrated that adiponectin participate in body weight regulation. Furthermore, in contrast to other adipokines such as leptin, serum level of adiponectin is negatively associated with insulin resistance, hyperinsulinemia and obesity (Arita et al., 1999, Yatagai et al., 2003).
Adiponectin, a circulating 244-amino acid protein, is mainly secreted from adipose tissue. It has been suggested that adiponectin has anti-diabetic, anti-inflammatory (Nishida et al., 2007), anti-angiogenic, and anti-proliferative effects (Fenton et al., 2008, Sugiyama et al., 2009). Adiponectin suppresses colonic epithelial proliferation and play a positive role in cell death (Brakenhielm et al. 2004). The effects of adiponectin on carcinogenesis are mediated by binding and activating its receptors, which are expressed in normal colonic epithelium and in colon cancer cells (Williams et al., 2008). Epidemiologic evidence has also demonstrated lower serum levels of adiponectin (Erarslan et al., 2009, Gonullu et al., 2010) in cases with CRC than controls. Furthermore, a positive association for leptin-adiponectin ratio and CRC risk has been demonstrated (Stocks et al., 2008). Finally, the potential associations between the adiponectin (ADIPOQ) (Kaklamani et al., 2008, Partida-Pérez et al., 2010, He et al., 2011, Al-Harithy and Al-Zahrani, 2012, Hu et al., 2013, Xu et al., 2013) and adiponectin receptor 1 (ADIPOR1) (Kaklamani et al., 2008, He et al., 2011, Liu et al., 2011) gene variants and CRC risk that have been investigated in several studies, have shown inconsistent results and the role of these genes in the etiology of CRC is still equivocal.
We therefore designed the present study to investigate the possible associations of ADIPOQ (rs2241766) and ADIPOR1 (rs2275738) gene variants with CRC risk.
2. Materials and methods
2.1. Participants
A total of 261 cases with CRC aged 22 to 84 years and 339 controls aged 19 to 85 years who referred to Taleghani Hospital (Tehran, Iran) between March 2008 and June 2012 were enrolled into the study. All the 600 subjects were Iranian and genetically unrelated. The case group consisted of all eligible colonoscopy patients with positive pathologic report for CRC, and eligibility criteria for the control group included no individual history of malignant colorectal tumors, adenomatous polyps, or other polyps. Before subjects' colonoscopy, interviews were performed by using a self-administered questionnaire and information regarding the subjects' demographic, anthropometric, and clinical characteristics was recorded. Prior entering the study, informed consent was obtained from each subject and the study was approved by the Ethical Committee of Gastroenterology and Liver Diseases Research Center, Shahid Beheshti University of Medical Sciences. Body mass index (BMI) of each subject was calculated as body weight divided by height squared (kg/m2) and the subjects were divided in the subgroups based on the diagnosis of CRC and BMI values as follows: normal weight (BMI < 25 kg/m2) controls (n = 166); overweight/obese (BMI ≥ 25 kg/m2) controls (n = 173); normal weight cases with CRC (n = 120); and overweight/obese cases with CRC (n = 141).
2.2. Genotype analysis
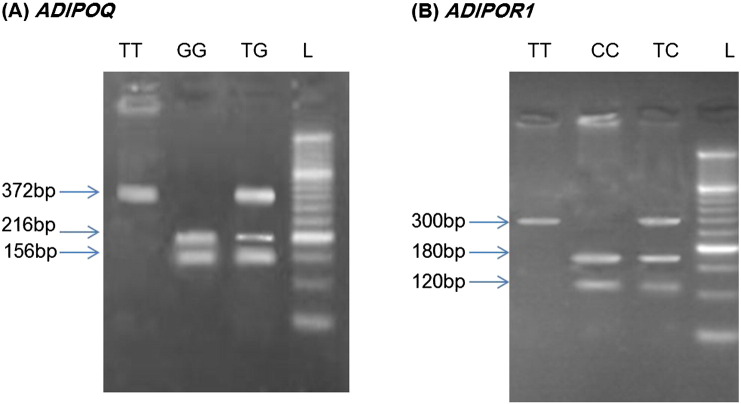
Five milliliters of peripheral blood samples from all 600 subjects were collected in tubes containing EDTA as an anticoagulant and store at 4 °C. Genomic DNA was isolated from peripheral blood leucocytes using standard methods, and genotyping was done by PCR-RFLP method. Characteristics of the studied gene variants, PCR primers, and PCR and RFLP conditions are summarized in Table 1. The two SNPs studied were selected based on their commonly use in previous genetic epidemiology studies, degree of heterozygosity, position in the gene, and functional importance. The PCR products were digested overnight with the appropriate restriction enzymes (Fermentas, Leon-Rot, Germany) and the digested products were run on 2 to 3% agarose gels (Fig. 1). Bands in gels stained with ethidium bromide for visualization under ultraviolet light. The concordance of genotyping was confirmed by duplicate analysis of approximately 10% of the randomly selected samples.
Table 1.
Information for the studied markers in adiponectin (ADIPOQ) and adiponectin receptor 1 (ADIPOR1) genes.
| Gene name (SNP ID) | Chromosome location (base change) | Forward primer Reverse primer |
PCR program (35 cycles) | PCR size (bp) | Restriction enzyme, incubation temperature | Alleles: RFLP size (bp) |
|---|---|---|---|---|---|---|
| ADIPOQ (rs2241766) | 3q27.3; Exon 2; (T > G) |
5′-GAAGTAGACTCTGCTGAGATGG-3′ 5′-TATCAGTGTAGGAGGTCTGTGATG-3′ |
93 °C 45 s,64 °C 30s, 72 °C 45 s |
372 |
SmaI, 30 °C |
Allele T: 372 Allele G: 216 + 156 |
| ADIPOR1 (rs2275738) | 1q32.1; Intron 1; (C > T) |
5′-TTTGTGGGAAGACTCTGGCTGGT-3′ 5′-TTAGTGAGGTTCTGGGTAAAGGTT GACATT-3′ |
93 °C 45 s,64 °C 30s, 72 °C 45 s |
300 |
RseI, 37 °C |
Allele T: 300 Allele C: 180 + 120 |
Fig. 1.
Representative results of the PCR-RFLP analysis for ADIPOQ (A) and ADIPOR1 (B) gene variants detected by agarose gel electrophoresis. Lane ‘L’ illustrates the 50 bp DNA ladder.
2.3. Statistical methods
Differences in demographic or anthropometric factors were calculated using t-test or chi-square test when appropriate. Testing Hardy-Weinberg equilibrium (HWE) for each of the two gene variants among cases and controls, separately, and comparisons of the distribution of the allele frequencies between the groups were performed using the chi-square test. Logistic regression was used to examine genotype frequencies between the different groups. We also used logistic regression analysis to adjust for confounding factors. All statistical analyses were conducted by using SPSS software (version 15.0; SPSS Inc. Chicago, IL, USA) and P-values < 0.05 were considered statistically significant (He et al., 2016a, He et al., 2016b).
3. Results
3.1. Clinicopathological analysis
Selected characteristics of the study population and their statistical significance are summarized in Table 2. In general, cases with CRC were older and less likely to use NSAIDs when compared with their control counterparts. However, there were no significant differences between the cases with CRC and the controls in terms of sex, BMI, smoking status, and family history of CRC.
Table 2.
Selected characteristics of colorectal cancer patients and control subjectsa.
| Variables | Controls (n = 339) |
Cases (n = 261) |
P-value |
|---|---|---|---|
| Age (years) | 44.3(16.3) | 56.1(12.6) | < 0.001 |
| BMI(kg/m2) | 25.2(4.2) | 25.6(4.9) | 0.261 |
| Gender | |||
| Men | 164(48.4) | 146(55.9) | |
| Women | 175(51.6) | 115(44.1) | 0.066 |
| Smoking history | |||
| No | 290(85.5) | 214(82.0) | |
| Former | 39(11.5) | 32(12.3) | |
| Current | 10(3.0) | 15(5.7) | 0.261 |
| Regular NSAID use | |||
| No | 270(79.6) | 248(95.0) | |
| Yes | 69(20.4) | 13(5.0) | < 0.001 |
| Family history of colorectal cancer | |||
| No | 303(89.4) | 229(87.7) | |
| Yes | 36(10.6) | 32(12.3) | 0.530 |
| Tumor site | |||
| Colon | – | 167(63.9) | |
| Rectal | – | 94(36.1) | – |
| Metastasis | |||
| No | – | 176(89.3) | |
| Yes | – | 21(10.7) | – |
Variables presented as mean (SD) or number (%).
3.2. ADIPOQ and ADIPOR1 SNPs analysis
The distribution of genotypes and alleles of the ADIPOQ (rs2241766) and ADIPOR1 (rs2275738) gene variants in cases with CRC and controls are provided in Table 3. None of the genotype frequency distributions for the ADIPOR1 gene rs2275738 variant deviated significantly from the Hardy-Weinberg equilibrium in both cases and controls (P > 0.05), suggesting that the alleles are in equilibrium. However, the genotype frequencies of the ADIPOQ gene rs2241766 variant were consist with HWE among the cases, but were out of HWE in controls (P < 0.05), with decreased heterozygosity.
Table 3.
Association between genotypes and alleles of adiponectin (ADIPOQ) and adiponectin receptor 1 (ADIPOR1) gene variants and risk of colorectal cancera.
| Gene (variant) | Controls (n = 339) | Cases (n = 261) | OR (95%CI) P-valueb |
|---|---|---|---|
| ADIPOQ (rs2241766) | |||
| Genotype-wise comparison | |||
| TT | 240(70.8) | 199(76.2) | 1.0(reference) |
| TG | 82(24.2) | 55(21.1) | 0.87(0.57–1.33)0.520 |
| GG | 17(5.0) | 7(2.7) | 0.44(0.17–1.14)0.092 |
| TG and GG | 99(29.2) | 62(23.8) | 0.79(0.53–1.17)0.241 |
| GG versus others | 17(5.0) | 7(2.7) | 0.45(0.18–1.17)0.103 |
| Allele-wise comparison | |||
| T | 562(82.9) | 453(86.8) | 1.0(reference) |
| G | 116(17.1) | 69(13.2) | 1.36(0.98–1.87)0.064 |
| ADIPOR1 (rs2275738) | |||
| Genotype-wise comparison | |||
| TT | 109(32.2) | 63(24.1) | 1.0(reference) |
| TC | 154(45.4) | 134(51.4) | 1.50(0.99–2.29)0.056 |
| CC | 76(22.4) | 64(24.5) | 1.45(0.89–2.37)0.139 |
| TC and CC | 230(67.8) | 198(75.9) | 1.49(1.00–2.20)0.048 |
| CC versus others | 76(22.4) | 64(24.5) | 1.12(0.74–1.70)0.579 |
| Allele-wise comparison | |||
| T | 372(54.9) | 260(49.8) | 1.0(reference) |
| C | 306(45.1) | 262(50.2) | 0.82(0.65–1.03)0.082 |
Boldface data indicates significance at P-value < 0.05.
Variables presented as number (%).
Adjusted for age, BMI, sex, smoking status, NSAID use, and family history in genotype-wise comparisons.
As shown in Table 3, no significant difference was observed in genotype and allele frequencies between the cases and controls for ADIPOQ gene either before or after adjustment for confounding factors. However, the ADIPOQ rs2241766 “G” allele compared with the “T” allele was significantly underrepresented in overweight/obese cases with CRC than overweight/obese controls (P = 0.046; OR = 0.64, 95%CI = 0.41–0.99). More importantly, analysis of the ADIPOR1 − 106 C > T (rs2275738) variant revealed a significant difference between cases with CRC and controls. These data indicated that the carriers of the ADIPOR1 “CC + CT” genotype compared with “TT” genotype occurred more frequently in cases with CRC than controls (P = 0.032; OR = 1.50, 95%CI = 1.04–2.14), and the difference remained significant after adjustment for age, BMI, sex, smoking status, NSAID use, and family history of CRC (P = 0.048; OR = 1.49, 95%CI = 1.01–2.20).
In this study the risk of obesity in relation to the two gene variants was also examined. Interestingly, these data indicated that the carriers of the ADIPOR1 rs2275738 “CC + TC” genotype compared with “TT” genotype occurred more frequently in overweight/obese cases with CRC than normal weight cases with CRC, and the difference remained significant after adjustment for age, sex, smoking status, NSAID use, and family history of CRC (P = 0.040; OR = 1.86, 95%CI = 1.03–3.36).
4. Discussion
We conducted a case-control study to explore the possible association between the ADIPOQ (rs2241766) and ADIPOR1 (rs2275738) gene variants and CRC risk among Iranians. No significant difference was found for ADIPOQ gene in either genotype or allele frequencies between the cases with CRC and controls. However, the ADIPOQ rs2241766 “G” allele compared with the “T” allele was significantly underrepresented in overweight/obese cases with CRC than overweight/obese controls. More importantly, our findings also indicated for the first time that the ADIPOR1 rs2275738 “CC + CT” genotype compared with “TT” genotype to be marker of increased CRC susceptibility and the difference remained significant after adjustment for the confounding factors. Furthermore, the ADIPOR1 “CC + TC” genotype compared with “TT” genotype was associated with an increased risk for obesity in the cases with CRC.
4.1. ADIPOQ gene rs2241766 variant
Complex diseases such as CRC are influenced by interacting networks of genetic and environmental factors. One method of identifying novel susceptibility genes for the diseases is to study polymorphisms in candidate genes and therefore the association among DNA sequence variations and CRC has become a subject of interest in recent years. Adipokine genes including ADIPOQ and ADIPOR1 are strong candidates for the link between obesity and risk of CRC. The potential association between the rs2241766 variant of ADIPOQ gene and CRC risk has been investigated in several studies. Some studies have reported no associations between the rs2241766 variant and the CRC risk (Kaklamani et al., 2008, Partida-Pérez et al., 2010, He et al., 2011). In our study, no significant difference was also found for the variant between the cases and controls. However, our findings in overweight/obese subjects are in line with three recent studies (Al-Harithy and Al-Zahrani, 2012, Hu et al., 2013, Xu et al., 2013) showing a significant association between the rs2241766 variant and CRC risk. We observed that the ADIPOQ rs2241766 “G” allele was associated with decreased risk of CRC in overweight/obese subjects (P = 0.046; OR = 0.64, 95%CI = 0.41–0.99). A very recent meta-analysis (Xu et al., 2013) indicated that the G allele was a protective factor against cancer, which is consistent with our results. Furthermore, Al-Harithy and Al-Zahrani (2012) observed that the ADIPOQ rs2241766 “GG + GT” genotype was associated with decreased risk of CRC; while Hu et al. (2013) found that the “GG + GT” genotype was related to the increased risk of CRC. The rs2241766 variant at exon 2 is a “synonymous” SNP meaning that it does not alter the amino acid sequence of adiponectin and therefore the exact molecular mechanism responsible for the relationship of the variant with CRC risk is not known. However, the rs2241766 variant may affect mRNA levels through regulation of mRNA splicing and/or stability. Yang et al. (2003) have suggested that the “G” allele of the rs2241766 variant appears to be more active and its expression was higher than the “T” allele. Previous studies have also reported lower serum levels of adiponectin (Erarslan et al., 2009, Gonullu et al., 2010) in cases with CRC than controls, and higher adiponectin levels in individuals with “GG” genotype compared with those with “TT” genotype (Heid et al., 2006). Accordingly, our finding that the ADIPOQ rs2241766 “G” allele appeared to be a marker of decreased CRC susceptibility in overweight/obese subjects is consistent with the notions above. However, in the present study the distribution of the ADIPOQ rs2241766 genotypes deviated from HWE in controls. Deviations from HWE may be attributable to genotyping error, population stratification, small sample size or a true association. Because our genotype data had high quality and all the individuals included in this study were Iranian, as their parents and grandparents were, it seems unlikely that genotyping error or population stratification results in departure from HWE. However, the deviation may be a function of small sample size, because the chance of a deviation from HWE decreases with increasing sample size. Another important point that we should keep in mind is that no departure from HWE would be observed in the control group if they were collected from the general population without any exclusion criteria. In other words, when individuals are excluded from control group due to not having the respective inclusion criteria, a violation of HWE will occur that may result in departure from HWE in the controls. Accordingly, the deviation from HWE can affect the interpretation of the association observed in this study, nonetheless, the possibility of a true association between ADIPOQ rs2241766 variant and CRC risk in overweight/obese subjects should not be excluded.
4.2. ADIPOR1 gene rs2275738 variant
Only three studies to date have evaluated the association between ADIPOR1 gene variants and CRC risk, and the results were contradictory (Kaklamani et al., 2008, He et al., 2011, Liu et al., 2011). However, to our knowledge, no studies have examined the association between the − 106 C > T (rs2275738) variant of ADIPOR1 gene and CRC risk up to now. In accord with the results obtained by Kaklamani et al. (2008) and He et al. (2011), we demonstrated a significant association between ADIPOR1 gene polymorphism and CRC risk. In contrast, in a study by Liu et al. (2011) no association was found. Our findings suggest for the first time that the ADIPOR1 − 106 C > T gene variant may be a genetic contributor to CRC risk. We found that the ADIPOR1 rs2275738 “CC + CT” genotype compared with the “TT” genotype was associated with an approximate 49% increased risk for CRC. In the study of Kaklamani et al. (2008), the presence of the ADIPOR1 rs1342387 “CC + CT” genotype compared with the TT genotype increased the risk for CRC, which was similar with the study of He et al. (2011). Interestingly, our study is also the first to show an association between the ADIPOR1 rs2275738 variant and obesity risk in the patients with CRC. We found that the ADIPOR1 rs2275738 “CC + CT” genotype compared with the “TT” genotype was associated with an approximate 86% increased risk for obesity. This finding is in concordance with the study of Siitonen et al. (2006), where the ADIPOR1 rs2275738 “CC” genotype was associated with the indicators of central obesity. The molecular mechanism through which the rs2275738 variant influences the risk of CRC and obesity is not known at present; however, previous studies have shown significant associations between the ADIPOR1 gene variants and serum levels of adiponectin (Heid et al., 2006) and BMI and insulin resistance (Siitonen et al., 2006). Previous studies have also demonstrated that the ADIPOR1 gene variants were associated with the regulation of ADIPOR1 gene expression (Soccio et al., 2006) and the expression of AdipoR1 is significantly higher in cancer tissue compared with non-tumor tissue from patients with CRC (Williams et al., 2008). The upregulation of AdipoR1 in tumor cells may be a cellular response to lower circulating adiponectin levels in patients with CRC. We may also infer that the rs2275738 variant could influence CRC and obesity risk by its linkage disequilibrium with other SNPs such as rs1342387 variant (Siitonen et al., 2006) to regulate the expression of ADIPOR1 gene, however, it remained to be confirmed. Accordingly, it is reasonable to believe that ADIPOR1 gene plays a role in pathogenesis of CRC.
4.3. Study limitations
Although well-designed, our study has several limitations. One limitation is the modest sample size that precludes drawing strong conclusions and doing detailed analyses. Another limitation is the limited number of polymorphisms examined. The other potential limitation is our lack of information on serum level of adiponectin as well as mRNA expression levels of ADIPOQ and ADIPOR1 genes, which could modify the effects observed here. However, this study certainly provides interesting information and may serve to guide future studies in this area.
4.4. Conclusions
To our knowledge, this study demonstrates for the first time that the − 106 C > T variant of ADIPOR1 gene may be a genetic contributor to CRC and obesity risk. This observation is relevant from a scientific standpoint; however, further large-scale studies will be needed to firmly establish the relationships between ADIPOR1 gene and CRC risk. Furthermore, the ADIPOQ rs2241766 “G” allele is a protective factor for CRC in overweight/obese subjects. However, further studies with bigger sample size are needed to validate these findings.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
The authors thank all patients and healthy blood donors for providing blood samples. This work was supported by a grant from Gastroenterology and Liver Diseases Research Center, Shahid Beheshti University of Medical Sciences (Grant number: 496).
References
- Al-Harithy R.N., Al-Zahrani M.H. The adiponectin gene, ADIPOQ, and genetic susceptibility to colon cancer. Oncol. Lett. 2012;3:176–180. doi: 10.3892/ol.2011.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita Y., Kihara S., Ouchi N., Takahashi M., Maeda K., Miyagawa J. Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem. Biophys. Res. Commun. 1999;257:79–83. doi: 10.1006/bbrc.1999.0255. [DOI] [PubMed] [Google Scholar]
- Brakenhielm E., Veitonmaki N., Cao R., Kihara S., Matsuzawa Y., Zhivotovsky B. Adiponectin-induced antiangiogenesis and anti tumor activity involve caspase-mediated endothelial cell apoptosis. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2476–2481. doi: 10.1073/pnas.0308671100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caan B.J., Coates A.O., Slattery M.L., Potter J.D., Quesenberry C.P., Edwards S.M. Body size and the risk of colon cancer in a large case-control study. Int. J. Obes. Relat. Metab. Disord. 1998;22:178–184. doi: 10.1038/sj.ijo.0800561. [DOI] [PubMed] [Google Scholar]
- Erarslan E., Turkay C., Koktener A., Koca C., Uz B., Bavbek N. Association of visceral fat accumulation and adiponectin levels with colorectal neoplasia. Dig. Dis. Sci. 2009;54:862–868. doi: 10.1007/s10620-008-0440-6. [DOI] [PubMed] [Google Scholar]
- Fenton J., Birmingham J., Hursting S., Hord N. Adiponectin blocks multiple signaling cascades associated with leptin-induced cell proliferation in Apc-Min/1 colon epithelial cells. Int. J. Cancer. 2008;122:2437–2445. doi: 10.1002/ijc.23436. [DOI] [PubMed] [Google Scholar]
- Ferlay J., Shin H.R., Bray F., Forman D., Mathers C., Parkin D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- Gonullu G., Kahraman H., Bedir A., Bektas A., Yücel I. Association between adiponectin, resistin, insulin resistance, and colorectal tumors. Int. J. Color. Dis. 2010;25:205–212. doi: 10.1007/s00384-009-0828-6. [DOI] [PubMed] [Google Scholar]
- He J., Yang T., Zhang R., Zhu J., Wang F., Zou Y. Potentially functional polymorphisms in the LIN28B gene contribute to neuroblastoma susceptibility in Chinese children. J. Cell Mol. Med. 2016 doi: 10.1111/jcmm.12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Zhong W., Zeng J., Zhu J., Zhang R., Wang F. LMO1 gene polymorphisms contribute to decreased neuroblastoma susceptibility in a Southern Chinese population. Oncotarget. 2016 doi: 10.18632/oncotarget.8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Pan Y., Zhang Y., Bao Q., Chen L., Nie Z. Effects of genetic variations in the adiponectin pathway genes on the risk of colorectal cancer in the Chinese population. BMC Med. Genet. 2011;12:94. doi: 10.1186/1471-2350-12-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid I.M., Wagner S.A., Gohlke H., Iglseder B., Muller J.C., Cip P. Genetic architecture of the APM1 gene and its influence on adiponectin plasma levels and parameters of the metabolic syndrome in 1,727 healthy Caucasians. Diabetes. 2006;55:375–384. doi: 10.2337/diabetes.55.02.06.db05-0747. [DOI] [PubMed] [Google Scholar]
- Hu X., Yuan P., Yan J., Feng F., Li X., Liu W. Gene polymorphisms of ADIPOQ + 45 T > G, UCP2 − 866 G > A, and FABP2 Ala54Thr on the risk of colorectal cancer: A matched case-control study. PLoS One. 2013;8 doi: 10.1371/journal.pone.0067275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaklamani V.G., Wisinski K.B., Sadim M., Gulden C., Do A., Offit K. Variants of the adiponectin (ADIPOQ) and adiponectin receptor 1 (ADIPOR1) genes and colorectal cancer risk. JAMA. 2008;300:1523–1531. doi: 10.1001/jama.300.13.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhong R., Wei S., Yin J.Y., Xiang H., Zou L. Interactions between genetic variants in the adiponectin, adiponectin receptor 1 and environmental factors on the risk of colorectal cancer. PLoS One. 2011;6 doi: 10.1371/journal.pone.0027301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida M., Funahashi T., Shimomura I. Pathophysiological significance of adiponectin. Med. Mol. Morphol. 2007;40:55–67. doi: 10.1007/s00795-007-0366-7. [DOI] [PubMed] [Google Scholar]
- Partida-Pérez M., de la Luz Ayala-Madrigal M., Peregrina-Sandoval J., Macías-Gómez N., Moreno-Ortiz J., Leal-Ugarte E. Association of LEP and ADIPOQ common variants with colorectal cancer in Mexican patients. Cancer Biomark. 2010;7:117–121. doi: 10.3233/CBM-2010-0154. [DOI] [PubMed] [Google Scholar]
- Schoen R.E., Tangen C.M., Kuller L.H., Burke G.L., Cushman M., Tracy R.P. Increased blood glucose and insulin, body size, and incident colorectal cancer. J. Natl. Cancer Inst. 1999;91:1147–1154. doi: 10.1093/jnci/91.13.1147. [DOI] [PubMed] [Google Scholar]
- Siitonen N., Pulkkinen L., Mager U., Lindstrom J., Eriksson J.G., Valle T.T. Association of sequence variations in the gene encoding adiponectin receptor 1 (ADIPOR1) with body size and insulin levels. The Finnish Diabetes Prevention Study. Diabetologia. 2006;49:1795–1805. doi: 10.1007/s00125-006-0291-7. [DOI] [PubMed] [Google Scholar]
- Soccio T., Zhang Y.Y., Bacci S., Mlynarski W., Placha G., Raggio G. Common haplotypes at the adiponectin receptor 1 (ADIPOR1) locus are associated with increased risk of coronary artery disease in type 2 diabetes. Diabetes. 2006;55:2763–2770. doi: 10.2337/db06-0613. [DOI] [PubMed] [Google Scholar]
- Stocks T., Lukanova A., Johansson M., Rinaldi S., Palmqvist R., Hallmans G. Components of the metabolic syndrome and colorectal cancer risk; a prospective study. Int. J. Obes. 2008;32:304–314. doi: 10.1038/sj.ijo.0803713. [DOI] [PubMed] [Google Scholar]
- Sugiyama M., Takahashi H., Hosono K., Endo H., Kato S., Yoneda K. ADIPOQ inhibits colorectal cancer cell growth through the AMPK/mTOR pathway. Int. J. Oncol. 2009;34:339–344. [PubMed] [Google Scholar]
- Trevisan M., Liu J., Muti P., Misciagna G., Menotti A., Fucci F. Risk factors and life expectancy research group. Markers of insulin resistance and colorectal cancer mortality. Cancer Epidemiol. Biomark. Prev. 2001;10:937–941. [PubMed] [Google Scholar]
- Williams C.J., Mitsiades N., Sozopoulos E., Hsi A., Wolk A., Nifli A.P. Adiponectin receptor expression is elevated in colorectal carcinomas but not in gastrointestinal stromal tumors. Endocr. Relat. Cancer. 2008;15:289–299. doi: 10.1677/ERC-07-0197. [DOI] [PubMed] [Google Scholar]
- Xu Y., He B., Pan Y., Gu L., Nie Z., Chen L. The roles of ADIPOQ genetic variations in cancer risk: evidence from published studies. Mol. Biol. Rep. 2013;40:1135–1144. doi: 10.1007/s11033-012-2154-2. [DOI] [PubMed] [Google Scholar]
- Yang W.S., Tsou P.L., Lee W.J., Tseng D.L., Chen C.L., Peng C.C. Allele-specific differential expression of common adiponectin gene polymorphism related to obesity. J. Mol. Med. 2003;81:428–434. doi: 10.1007/s00109-002-0409-4. [DOI] [PubMed] [Google Scholar]
- Yatagai T., Nagasaka S., Taniguchi A., Fukushima M., Nakamura T., Kuroe A. Hypoadiponectinemia is associated with visceral fat accumulation and insulin resistance in Japanese men with type 2 diabetes mellitus. Metabolism. 2003;52:1274–1278. doi: 10.1016/s0026-0495(03)00195-1. [DOI] [PubMed] [Google Scholar]