Table 1. Precision and gene overlap for various competing models for the task of phenotype discovery, evaluated using disease-gene associations.
| Data | K | Average precision@K |
Average perc. of overlapping genes@K |
||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 5 | 10 | 1 | 2 | 5 | 10 | ||
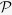 |
D2D | 0.9449 | 0.9367 | 0.9225 | 0.9047 | 0.8159 | 0.7966 | 0.7564 | 0.7111 |
| Modularity (Adjacency) | 0.8575 | 0.8457 | 0.8284 | 0.8130 | 0.5198 | 0.4893 | 0.4508 | 0.4145 | |
| Spectral (Adjacency) | 0.7181 | 0.7007 | 0.6795 | 0.6640 | 0.3052 | 0.2779 | 0.2311 | 0.2006 | |
| Modularity (Comorbidity) | 0.8493 | 0.8412 | 0.8110 | 0.7865 | 0.5586 | 0.5315 | 0.4681 | 0.4204 | |
| Spectral (Comorbidity) | 0.8375 | 0.8316 | 0.8190 | 0.7974 | 0.5288 | 0.4989 | 0.4520 | 0.3964 | |
| Comorbidity graph | 0.7268 | 0.7134 | 0.6915 | 0.7068 | 0.1582 | 0.1496 | 0.1465 | 0.1554 | |
| Disease co-occurrence | 0.5616 | 0.5516 | 0.5439 | 0.5668 | 0.1448 | 0.1329 | 0.1264 | 0.1261 | |
| LDA | 0.5260 | 0.5094 | 0.4913 | 0.4217 | 0.1040 | 0.0989 | 0.0864 | 0.0853 | |
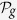 |
DAG2D | 0.9598 | 0.9444 | 0.9239 | 0.9079 | 0.8486 | 0.7963 | 0.7237 | 0.6720 |
| Modularity (Adjacency) | 0.8711 | 0.8604 | 0.8503 | 0.8389 | 0.5303 | 0.5082 | 0.4706 | 0.4340 | |
| Spectral (Adjacency) | 0.9165 | 0.9102 | 0.9020 | 0.8926 | 0.7524 | 0.7430 | 0.7277 | 0.7110 | |
| Disease and genes co-occurrence | 0.6978 | 0.6985 | 0.7093 | 0.7071 | 0.1058 | 0.1042 | 0.1018 | 0.0935 | |
| LDA | 0.5795 | 0.3874 | 0.3253 | 0.2831 | 0.1136 | 0.0781 | 0.0760 | 0.0652 | |
