Figure 4.
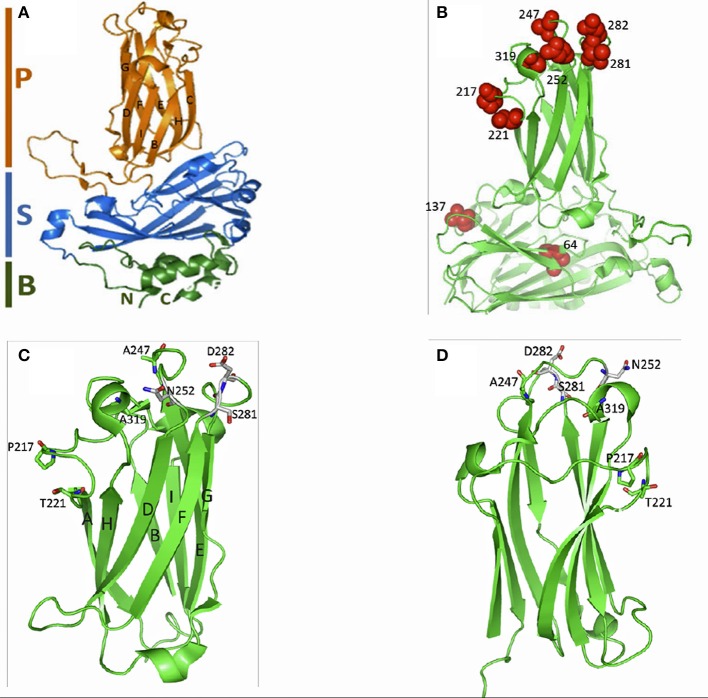
Subviral particle (SVP) of the VP2 capsid for IPNV. (A) Shows the SVP divided into the P-domain (magenta) made of the β-sheet that supports the surface loops that form the hypervariable region (HVR), S-domain (blue) that form the shell of the capsid, and the B-domain (green) that form of the base made of the N- and C-termini. (B) Shows the position of the residues linked to clinical and subclinical forms of IPNV infection (red balls). Note that positions V64I and A137T are located in the B-domain while T217P and A221T on loop PBC, residues T247A and V252N on PDE, residues T281S and N282D on PFG and A319E on PHI in the surface loops of the HVR in the P-domain. (C) Shows the P-domain having the β-sheets supporting the surface loops of the HVR. The structural layout of residues T217, A221T, T247, N252, S281, D282 and A319 are shown as sticks projecting out of the surface loops of the HVR. (D) Shows the P-domain at 1800 turn of (C) All figures were generated in Pymol v99 (PyMol, 2012) using the 3IDE template (Coulibaly et al., 2010) on the SWISS model workspace (Arnold et al., 2006).