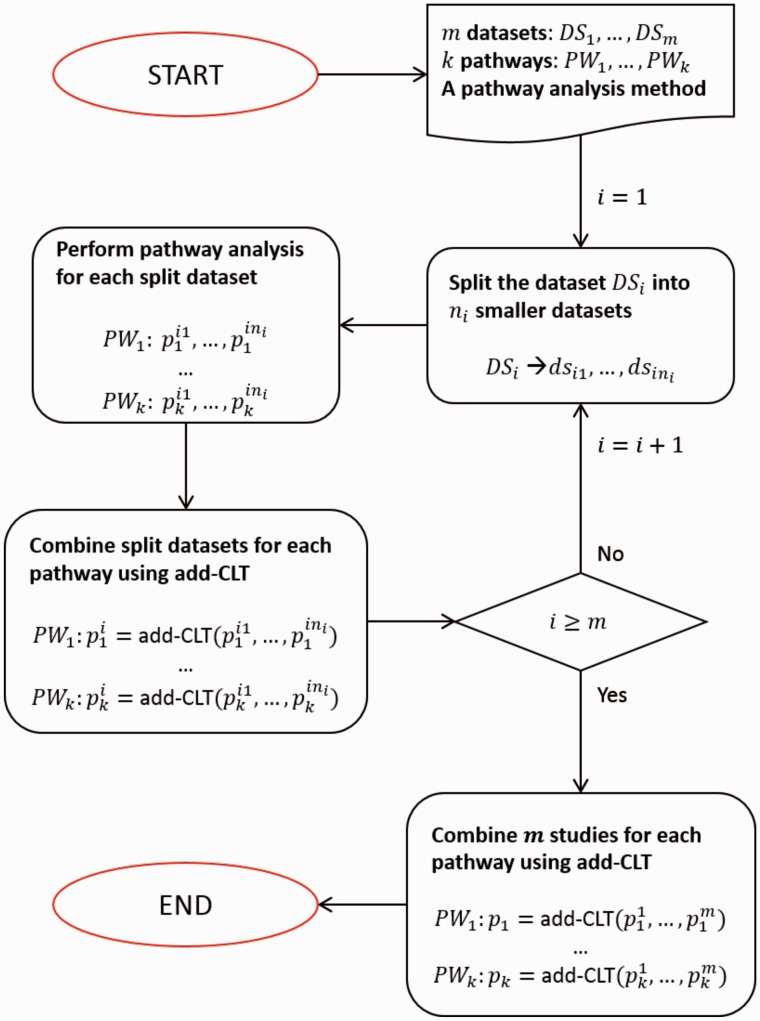
Fig. 2.
Bi-level meta-analysis framework to identify significant pathways. The input includes m datasets, k pathways and a pathway analysis method. The intra-experiment analysis divides the dataset DSi () into smaller datasets and then performs pathway analysis for each of the small datasets, resulting in ni P-values for each pathway. The intra-experiment analysis combines the ni P-values into one P-value for each pathway using add-CLT. After this process is done for all m studies (datasets), each pathway has m independent P-values – one per study. The inter-experiment analysis then combines the m P-values for each pathway into one meta P-value using the add-CLT method. This meta P-value for each pathway represents the overall significance of the pathway. The output of the framework is a list of k pathways ranked according to the meta P-values