Fig. 1.
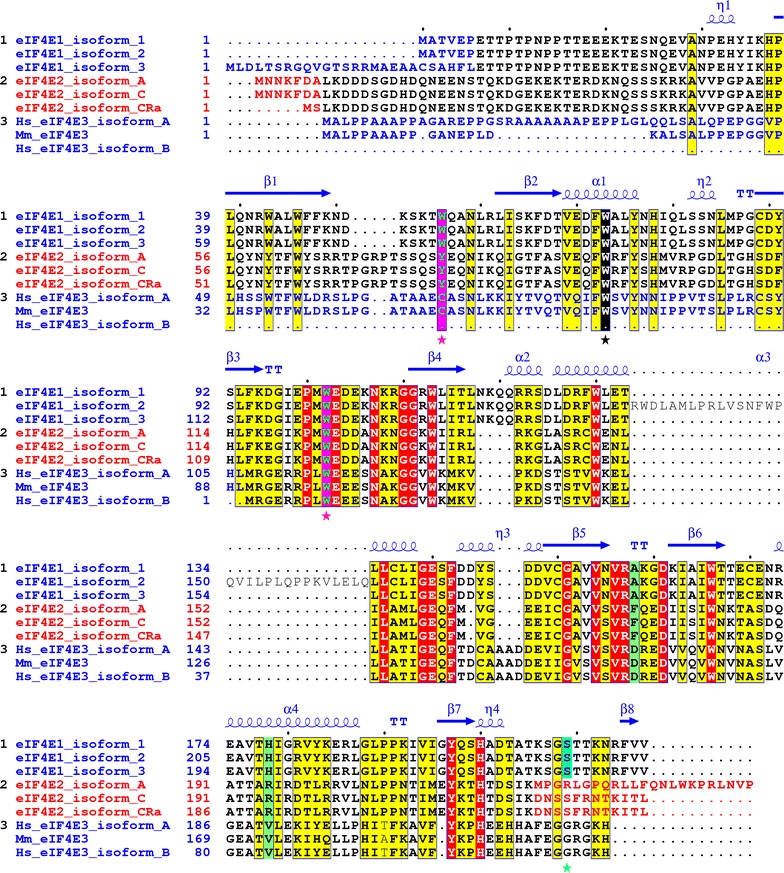
Alignment of the human eIF4E1, eIF4E2 and eIF4E3 isoforms and their variants explored in this study. Yellow and red boxes denote amino acids high similarity and identity, respectively. Utilization of alternative exons coding for different N- and C-protein termini is marked with blue and red letters. Cap-binding residues W56 and W102 in eIF4E1 and corresponding amino acids in eIF4E2 and eIF4E3 are highlighted as green letters in purple boxes and marked with purple asterisks. The conserved W73 (on the basis of eIF4E1_1) is marked with black box and black asterisk. Ser209 in eIF4E1 (numbering as of eIF4E1_1) is shaded in turquoise blue. Mouse eIF4E3 was added to highlight differences in primary structure between mouse and human orthologs. PDB file 3AM7 was used to depict eIF4E1 secondary structure [56]
