Fig. 1.
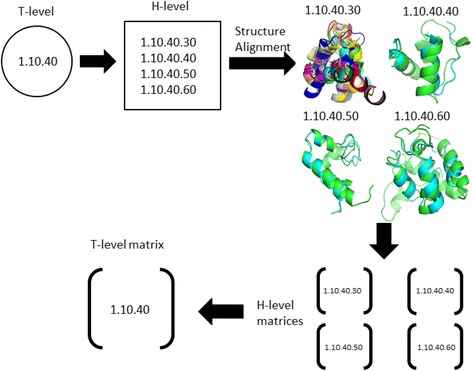
Example of how the amino acid similarity matrices are developed for one topology T-level (1.10.40). The available homology level (H-level) structure alignments are first used to obtain homology level similarity matrices. Then these homology level matrices (1.10.40.30, 1.10.40.40, 1.10.40.50, 1.10.40.60) are added together to yield the corresponding topology level matrix (1.10.40). Further decomposition of topology levels into homology level is required because homology level structural similarity is necessary to obtain reasonable structure alignments.
