Fig. 5.
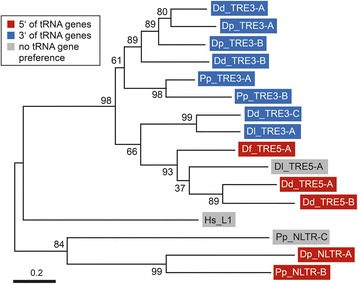
Phylogenetic analysis of dictyostelid non-LTR retrotransposons. Alignment of RT domains was generated with ClustalX and analyzed using the Maximum Likelihood method. All positions containing gaps and missing data were eliminated. The tree is drawn to scale with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. There were a total of 227 amino acid positions in the final dataset. Bootstrap support (percentage from 1000 trials) is indicated next to each node
