Fig. 6.
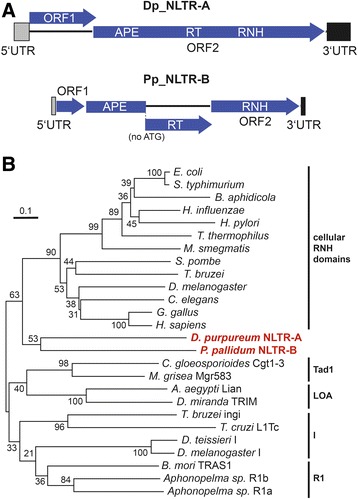
Phylogenetic analysis of RNH domains in dictyostelid NLTR elements. a Schematic presentations of the structures of D. purpureum NLTR-A and P. pallidum NLTR-B. See Fig. 1 for abbreviations. b Sequences of RNH domains were analyzed using the Neighbor Joining method. All positions containing gaps and missing data were eliminated. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Numbers next to each node indicate bootstrap values as percentages out of 1000 replicates [68]. Analysis of the data with the Maximum Likelihood method produced the same tree topology with slightly lower bootstrap values. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The tree was rooted on cellular RNH domains. Sequences used for alignment with D. purpureum NLTR-A and P. pallidum NLTR-B were chosen according to a previous phylogenetic analysis by performed by Malik et al. [11]: Drosophila miranda TRIM (X59239), Aedes aegypti Lian (U87543), Colletotrichum gloeosporioides Cgt1-3 (L76169), Magnaporthe grisea Mgr583 (AF018033), Trypanosoma bruzei ingi (X05710). Trypanosoma cruzi L1Tc (X83098), Drosophila teissieri I (M28878), Drosophila melanogaster I (M14954), Bombyx mori TRAS (GenBank D38414), and Aphonopelma sp. R1a (AF015489) and R1b (AAB94039). Cellular RNH domains used as outgroups were follows: Escherichia coli (P00647), Salmonella typhimurium (P23329), Buchnera aphidicola (Q08885), Haemophilus influenzae (P43807), Helicobacter pylori (P56120), Thermus thermophilus (P29253), Mycobacterium smegmatis (Q07705), Schizosaccharomyces pombe (AAC04366). Trypanosoma bruzei (AAC47537), Drosophila melanogaster (AAC47810), Caenorhabditis elegans (AAA83453). Gallus gallus (BAA05382), and Homo sapiens (CAA11835)
