Figure 3.
The X-Ray Structure of Pf-Avr4 Solved to 1.7-Å Resolution.
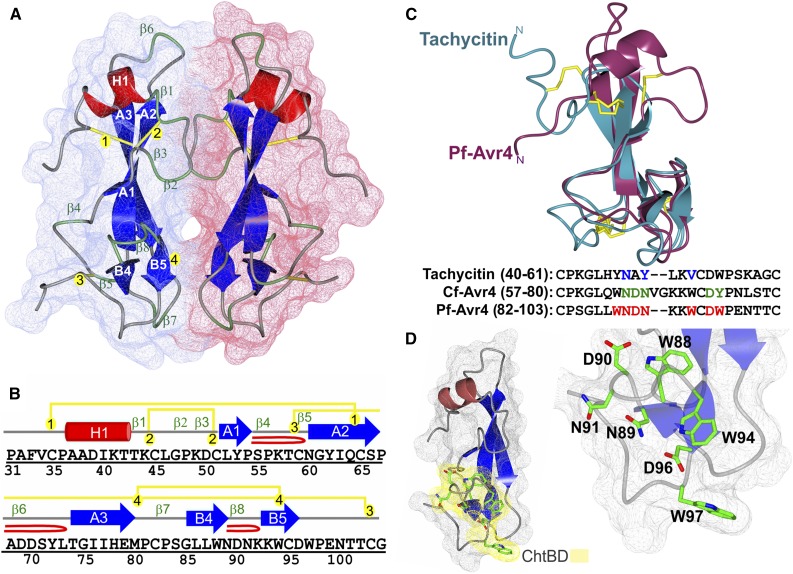
(A) The overall fold and surface filling of Pf-Avr4, spanning residues Pro-31 to Gly-105. Pf-Avr4 was crystallized with two molecules per asymmetric unit (chain A, red; B, blue), which are held together by many indirect water-mediated contacts (not shown) and three hydrogen-bond direct protein-protein contacts (not shown). The structure of Pf-Avr4 reveals a compact, globular protein stabilized by four disulfide bonds and an extensive network of intramolecular forces. Pf-Avr4 consists of a single N-terminal helix (red, H1) followed by a distorted β-sandwich fold, comprised of two β-sheets, one three-stranded, and the other two-stranded. The five β-strands are depicted in blue, labeled by their sheet (A and B) and placement within the structure (1 to 5). β-Turns are labeled in green (β1-8), and disulfide bonds are drawn in yellow and labeled by order in the structure (1 to 4).
(B) The secondary structure sequence alignment for Pf-Avr4, residues Pro-31 to Gly-105. Secondary structure elements are color-coded and labeled as depicted in (A). β-Hairpins are depicted by a red loop.
(C) Pf-Avr4 (maroon) aligns to CBM14 family member tachycitin (cyan) (RMSD 1.98 Å, for 52 aligned α-carbons), encompassing the distorted β-sandwich motif and putative ChtBD of tachycitin. Sequence alignment of tachycitin, Cf-Avr4, and Pf-Avr4 through their respective putative ChtBDs enabled the identification of seven residues (red, bold) on Pf-Avr4 that may have a key role in Pf-Avr4-chitin binding. Putative functional residues are shown for tachycitin (blue) and Cf-Avr4 (green).
(D) Pf-Avr4’s ChtBD (yellow) is located at the C terminus, and individual ChtBD residues (green) are illustrated and labeled on the ChtBD-magnified structure on the right.