Microdomains rich in 2-hydroxy sphingolipid play an essential role in chitin-triggered immunity by stimulating ROS production through the Rac/ROP small GTPase Rac1 and NADPH oxidases RbohB/H in rice.
Abstract
Numerous plant defense-related proteins are thought to congregate in plasma membrane microdomains, which consist mainly of sphingolipids and sterols. However, the extent to which microdomains contribute to defense responses in plants is unclear. To elucidate the relationship between microdomains and innate immunity in rice (Oryza sativa), we established lines in which the levels of sphingolipids containing 2-hydroxy fatty acids were decreased by knocking down two genes encoding fatty acid 2-hydroxylases (FAH1 and FAH2) and demonstrated that microdomains were less abundant in these lines. By testing these lines in a pathogen infection assay, we revealed that microdomains play an important role in the resistance to rice blast fungus infection. To illuminate the mechanism by which microdomains regulate immunity, we evaluated changes in protein composition, revealing that microdomains are required for the dynamics of the Rac/ROP small GTPase Rac1 and respiratory burst oxidase homologs (Rbohs) in response to chitin elicitor. Furthermore, FAHs are essential for the production of reactive oxygen species (ROS) after chitin treatment. Together with the observation that RbohB, a defense-related NADPH oxidase that interacts with Rac1, is localized in microdomains, our data indicate that microdomains are required for chitin-induced immunity through ROS signaling mediated by the Rac1-RbohB pathway.
INTRODUCTION
Plants have developed sophisticated immune systems to protect themselves from pathogen attack (Dodds and Rathjen, 2010). The first layer of defense responses is initiated by the detection of microbe-associated molecular patterns (MAMPs) by host pattern recognition receptors. This immune system is termed MAMP-triggered immunity (MTI) and leads to the generation of reactive oxygen species (ROS), calcium influx, and biosynthesis of phytoalexins (Boller and Felix, 2009). The second layer is effector-triggered immunity (ETI), which is induced by the perception of pathogen effectors by resistance (R) proteins (Cui et al., 2015). In both immune systems, the plasma membrane (PM) is important because it is the first point of contact between plant cells and pathogens. Many proteins that are crucial for immunity are therefore associated with the PM, including most pattern recognition receptors in MTI and several R proteins such as RPM1 and Pit in ETI (Antolín-Llovera et al., 2012; Gao et al., 2011; Kawano et al., 2010). In rice (Oryza sativa), the Rac/ROP small G protein Rac1, which acts as a molecular switch in defense signaling, is anchored to the PM by lipid modification, and respiratory burst oxidase homologs (Rbohs), which are plant NADPH oxidases that produce ROS, span the PM with six transmembrane domains (Kawano and Shimamoto, 2013; Kawano et al., 2014a; Marino et al., 2012). Effective regulation of PM-localized proteins is thus necessary for an early response to pathogen infection in plants.
Recent reports have suggested that specific proteins localize in PM microdomains (or membrane rafts), which are small, heterogeneous, liquid-ordered (Lo) domains composed mainly of sphingolipids and sterols, unlike the surrounding liquid-disordered (Ld) membrane composed of unsaturated glycerolipids (Mongrand et al., 2010; Simon-Plas et al., 2011). Plant PM microdomains contain a large number of proteins, including defense-related proteins (Mongrand et al., 2004; Morel et al., 2006; Kierszniowska et al., 2009). In addition, various specific proteins such as receptor-like kinases and dynamin-related proteins accumulate in microdomains after elicitation by, for example, chitin, cryptogein, and flagellin (Fujiwara et al., 2009; Stanislas et al., 2009; Keinath et al., 2010). Therefore, while PM microdomains are considered likely to be important for innate immunity in plants, it remains unknown whether they are actually necessary for the regulation of innate immunity. In other words, the biological significance of the existence of defense-related proteins in PM microdomains is unclear.
Sphingolipids are a large family of lipids that are ubiquitously present in eukaryotes and enriched in PM microdomains (Cacas et al., 2012). In plants, ceramide (Cer) is composed of a long-chain base (LCB) and a fatty acid, and the addition of various head groups including sugars or phosphates to Cers generates complex sphingolipids such as glucosylceramide (GlcCer) and glycosylinositolphosphoceramide (GIPC) (Supplemental Figure 1). Of the hundreds of sphingolipid species that exist in plants as a result of such modifications, many possess 2-hydroxy fatty acids (2-HFAs), containing a hydroxylated C-2 position (Markham et al., 2013) (Supplemental Figure 1A). 2-HFA contributes to the rigid binding of sphingolipids through hydrogen bonding between hydroxy groups in artificial membranes (Pascher and Sundell, 1977; Löfgren and Pascher, 1977), and sphingolipids containing 2-HFAs (2-hydroxy sphingolipids) reduce the lateral mobility of adipocyte membranes (Guo et al., 2010). Therefore, it is possible that 2-hydroxy sphingolipids contribute to the ordered structure of PM microdomains.
We have shown that 2-hydroxylation of sphingolipid fatty acids in Arabidopsis thaliana is performed by the enzyme fatty acid 2-hydroxylase (FAH) (Nagano et al., 2009, 2012a). Both Arabidopsis FAHs (At-FAH1 and At-FAH2) are localized to the endoplasmic reticulum (ER) membrane, and decreasing their levels affects the response to biotic and abiotic stresses (Nagano et al., 2009, 2012a; König et al., 2012). Thus, we hypothesized that FAH could be a useful tool to modify PM microdomains and illuminate the relationship between PM microdomains and innate immunity in plants.
In this study, we first confirmed the effect of 2-hydroxy sphingolipids on the formation of PM microdomains in vivo, using rice lines in which 2-hydroxy sphingolipids were decreased by knockdown of endogenous FAH homologs. We then demonstrated the importance of PM microdomains in disease resistance to rice blast fungus. Furthermore, analyses of defense-related PM microdomain proteins suggested a mechanism by which PM microdomains regulate rice innate immunity.
RESULTS
Os-FAH1 and Os-FAH2 Catalyze the 2-Hydroxylation of Sphingolipid Fatty Acids
Rice possesses two FAH homologs (Os-FAH1 and Os-FAH2), which are highly similar to their Arabidopsis counterparts (Nagano et al., 2012b). However, since it is unknown whether rice FAHs function as sphingolipid fatty acid 2-hydroxylases, we first characterized these Os-FAHs. Experiments with RFP-fused Os-FAH1 and Os-FAH2 revealed that both proteins colocalized with an ER-associated CFP reporter (Supplemental Figure 2). We generated transgenic FAH1/FAH2 double-knockdown lines (OsFAH1/2-KD) using an RNA interference (RNAi) system and confirmed that they grew more slowly than the wild type in suspension cells and looked smaller in plants (Supplemental Figure 3).
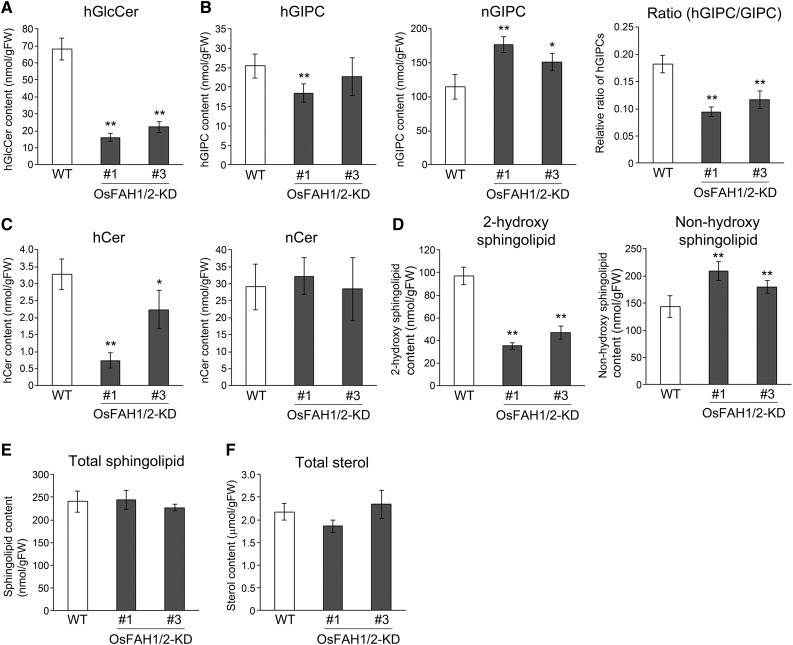
To examine the involvement of Os-FAHs in the 2-hydroxylation of sphingolipid fatty acids, we analyzed the sphingolipids containing 2-HFAs or non-hydroxy fatty acids (NFAs) with 20, 22, or 24 carbons (GlcCers, GIPCs, and Cers) (Supplemental Figure 1), extracted from whole cells of wild-type and OsFAH1/2-KD suspension cells, by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Figure 1; Supplemental Figures 4 and 5). GlcCers containing 2-HFAs (hGlcCers) clearly decreased in OsFAH1/2-KD lines compared with the wild type (Figure 1A), whereas GlcCers containing NFAs (nGlcCers) were not detected because of their low amounts in rice. By contrast, the amount of GIPCs containing 2-HFAs (hGIPCs) did not decrease in OsFAH1/2-KD3 due to increasing amounts of h24:0 (Figure 1B; Supplemental Figures 4A and 5B). However, the ratio of hGIPCs to total GIPCs decreased significantly in both OsFAH1/2-KD lines because GIPCs containing NFAs (nGIPCs) increased (Figure 1B; Supplemental Figures 4B and 5B). The amount of Cers containing 2-HFAs (hCers) was lower in OsFAH1/2-KD lines than in the wild type, whereas the amount of Cers containing NFAs (nCers) was equal (Figure 1C). Thus, although the h24:0 level in total sphingolipids was not altered (Supplemental Figure 5D), total 2-hydroxy sphingolipids decreased markedly in OsFAH1/2-KD lines (Figure 1D). By contrast, total sphingolipid content in OsFAH1/2-KD lines was maintained at the same level as in the wild type because sphingolipids containing NFAs (non-hydroxy sphingolipids) increased (Figures 1D and 1E). These results indicate that OsFAHs 2-hydroxylate sphingolipid fatty acids in rice. On the other hand, the amount of sterols, which are other major components of PM microdomains, was similar in whole cells (Figure 1F).
Figure 1.
Sphingolipid and Sterol Content in OsFAH1/2-KD Lines.
Total lipids were extracted from wild-type and OsFAH1/2-KD suspension cells, and the indicated lipids were quantified by LC-MS/MS. Data are means ± sd (n = 4). Asterisks indicate significant differences compared with the wild type (Student’s t test; *P < 0.05, **P < 0.01).
(A) to (D) Amounts of each sphingolipid class ([A], GlcCer; [B], GIPC; [C], Cer; [D], total sphingolipid) are shown for 2-HFA- (“h” prefix) and NFA-containing (“n”) types; no GlcCer-containing NFA was detected in this study. For GIPC, the ratio of hGIPC to total GIPC is also shown.
(E) Total sphingolipid content, comprising both 2-hydroxy sphingolipids and non-hydroxy sphingolipids.
(F) Amount of total sterols.
2-Hydroxy Sphingolipids Build PM Microdomains
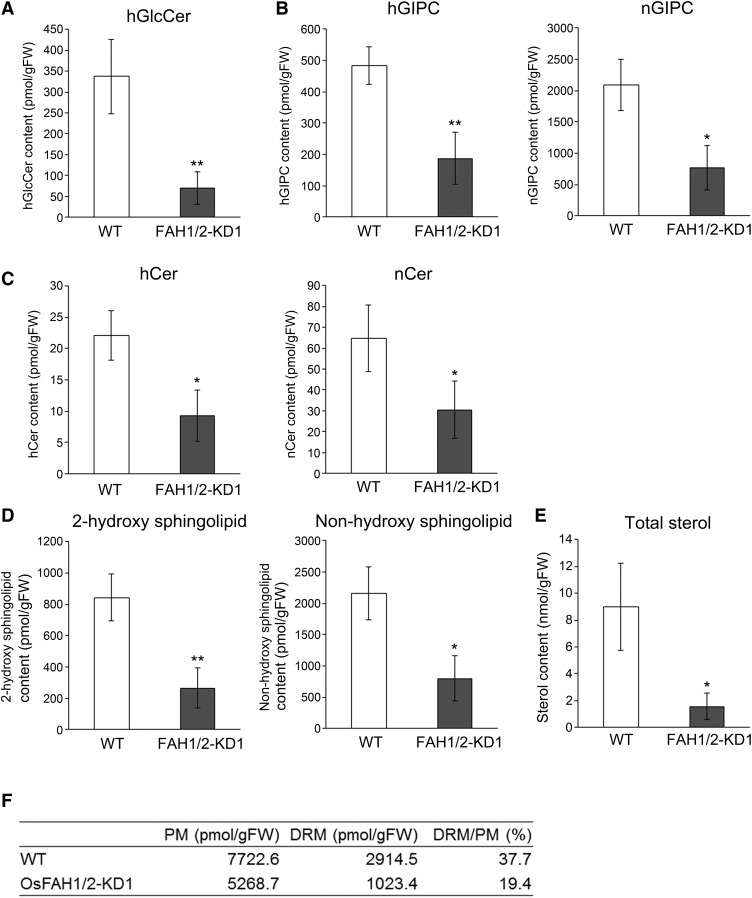
To elucidate whether 2-hydroxy sphingolipids are involved in the formation of PM microdomains in rice, we analyzed the sphingolipid and sterol level of the detergent-resistant membrane (DRM), which has often been used in analyses of lipids and proteins in microdomains, although they may include artifacts introduced by detergent treatment (Lingwood and Simons, 2007; Takahashi et al., 2013). PM was isolated from wild-type and OsFAH1/2-KD suspension cells by a two-phase partition system, and the DRM fraction was purified by OptiPrep density gradient centrifugation after Triton X-100 treatment. The amounts of both 2-hydroxy sphingolipids and non-hydroxy sphingolipids were significantly lower in the DRM of OsFAH1/2-KD1 than of the wild type (Figures 2A to 2D; Supplemental Figure 6), and sterol content also decreased in DRMs of OsFAH1/2-KD1 (Figure 2E). By contrast, non-hydroxy sphingolipid content in PM of OsFAH1/2-KD decreased less than in DRM (Supplemental Figures 7 and 8), and the ratio of DRM to PM decreased in OsFAH1/2-KD1 (Figure 2F). These results suggest that the amount of DRM extracted from PM is lower in OsFAH1/2-KD1.
Figure 2.
Lipid Analysis in the DRM Fractions of OsFAH1/2-KD1.
(A) to (D) Amounts of each sphingolipid class ([A], GlcCer; [B], GIPC; [C], Cer; [D], total sphingolipid) in DRMs extracted from PM of wild-type and OsFAH1/2-KD1 suspension cells are shown for 2-HFA- and NFA-containing types. Data are means ± sd (n = 4). Asterisks indicate significant differences compared with the wild type (Student’s t test; *P < 0.05, **P < 0.01).
(E) Amount of total sterols in DRMs extracted from PM of wild-type and OsFAH1/2-KD1 suspension cells. Data are means ± sd (n = 4). Asterisk indicates a significant difference compared with the wild type (Student’s t test; *P < 0.05).
(F) Amounts of PM and DRM and the ratio of DRM to PM in the wild type and OsFAH1/2-KD. Each amount indicates the sum of GlcCers and GIPCs.
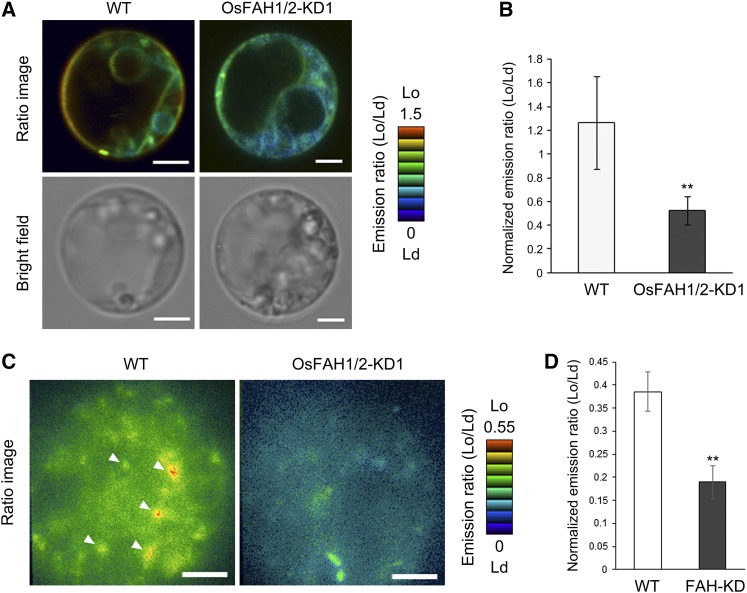
To further examine the effects of 2-hydroxy sphingolipids on the organization of PM microdomains, we visualized the PM phase of OsFAH1/2-KD1 in vivo using di-4-ANEPPDHQ, a dye that can distinguish between Lo and Ld phases in living cells (Owen et al., 2011). When we stained protoplasts from rice suspension cells with di-4-ANEPPDHQ, excited them at 488 nm, and simultaneously measured the emission ratio of Lo fluorescence (500 to 550 nm) to Ld fluorescence (650 to 750 nm) by confocal laser scanning microscopy (CLSM), the wild-type PM was more ordered than endomembranes (Figure 3A, left). However, the PM phase in OsFAH1/2-KD1 was significantly more disordered than in the wild type (Figures 3A and 3B). We investigated the PM phase in greater detail by total internal reflection fluorescence microscopy (TIRFM), a technique that is capable of detecting the fluorescence and dynamics of individual PM protein molecules (Fujimoto et al., 2007). As shown in Figures 3C and 3D, a large number of highly ordered puncta with a diameter of 246 ± 94 nm (n = 75) was observed in the PM of wild-type cells, whereas the heterogeneity of PM order in OsFAH1/2-KD was strikingly lower. PM microdomains are highly ordered structures in the PM (Mongrand et al., 2010; Simon-Plas et al., 2011), suggesting that the highly ordered puncta observed in this study corresponded to PM microdomains. Together with the results of DRM lipid analysis, these data indicate that 2-hydroxy sphingolipids play a key role in the formation of PM microdomains in rice cells.
Figure 3.
Observation of Membrane Order of OsFAH1/2-KD1.
(A) Membrane order in wild-type and OsFAH1/2-KD1 suspension cells was visualized by di-4-ANEPPDHQ. Stained protoplasts were observed by CLSM. In the ratio images, eight colors from red to blue represent the observed range of Lo/Ld ratios. Bars = 3 μm.
(B) Quantification of normalized Lo/Ld emission ratios of PM. Data are means ± sd (n = 20). Asterisks indicate a significant difference compared with the wild type (Student’s t test; **P < 0.01).
(C) PM order in rice protoplasts from wild-type and OsFAH1/2-KD1 suspension cells stained by di-4-ANEPPDHQ and observed by TIRFM. Eight colors from red to blue represent the observed range of Lo/Ld ratios. Arrowheads indicate highly ordered puncta. Bars = 3 μm.
(D) Quantification of normalized Lo/Ld emission ratios of puncta. Data are means ± sd (n = 20). Asterisks indicate a significant difference compared with the wild type (Student’s t test; **P < 0.01).
Os-FAHs Contribute to Resistance to Rice Blast Fungus Infection
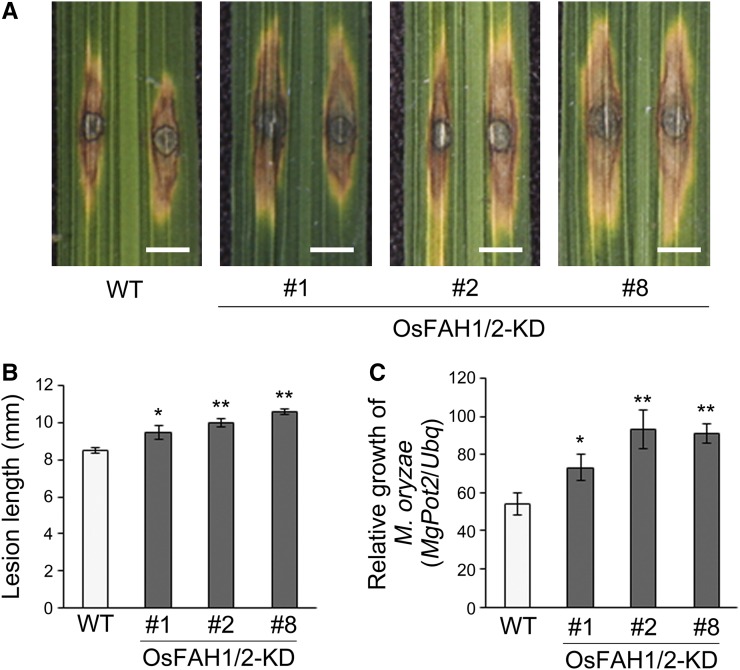
The above data indicated that sphingolipid- and sterol-dependent PM microdomains are constitutively reduced in OsFAH1/2-KD rice lines. We therefore used these knockdown lines to investigate the physiological role of PM microdomains in innate immunity by conducting a pathogen infection assay in OsFAH1/2-KD rice plants. When we inoculated similarly sized leaves of wild-type and OsFAH1/2-KD plants with compatible rice blast fungus (Magnaporthe oryzae strain 2403-1, race007.0), lesion length in OsFAH1/2-KD lines was significantly greater than in the wild type, and fungal growth was greater than in the wild type (Figure 4). This result suggests that PM microdomains produced by OsFAHs participate in the MTI pathway in rice. On the other hand, the salicylic acid (SA) level in OsFAH1/2-KD was similar to that in the wild type (Supplemental Figure 9), although the SA content increased in the Arabidopsis fah1 fah2 mutants described by König et al. (2012).
Figure 4.
Infection Assay for Rice Blast Fungus in OsFAH1/2-KD Plants.
(A) Infection of wild-type and OsFAH1/2-KD plants with a compatible strain of rice blast fungus (race007.0). Bars = 3 mm.
(B) Lesion length at 6 d postinfection in leaves of wild-type and OsFAH1/2-KD plants infected by the blast fungus. Data are means ± sd (n ≥ 38). Asterisks indicate significant differences compared with the wild type (Student’s t test; *P < 0.05, **P < 0.01).
(C) Relative growth of M. oryzae on susceptible rice cultivars. Data are means ± sd (n = 6). Asterisks indicate significant differences compared with the wild type (Student’s t test; *P < 0.05, **P < 0.01).
Numerous Defense-Related Proteins Are Decreased in OsFAH1/2-KD DRMs
To determine how Os-FAHs regulate innate immunity through PM microdomains in rice, we searched for PM microdomain proteins that are decreased in OsFAH1/2-KD cells by comparative proteome analysis of the DRM fractions of wild-type and OsFAH1/2-KD1 suspension cells. When total proteins of each DRM fraction were analyzed by LC-MS/MS, 314 proteins were detected in wild-type and 253 in OsFAH1/2-KD1 suspension cells (Supplemental Data Sets 1 and 2). By comparing the amounts of these proteins using the exponentially modified protein abundance index, which provides approximate relative quantitation of proteins in a mixture based on peptide matches in a database search result (Ishihama et al., 2005), we obtained 182 proteins whose abundance in OsFAH1/2-KD1 was <50% of that in the wild type (wild type > KD proteins; Table 1; Supplemental Data Set 3). This group included proteins such as transporters, cytoskeleton components, and membrane traffic-related proteins (Table 1; Supplemental Data Set 3). In agreement with the finding that OsFAH1/2-KD plants were more susceptible than the wild type to rice blast fungus, 35 candidates for defense-related proteins appeared in the wild type > KD group, including receptor-like kinases, leucine-rich repeat-containing proteins, chaperones, Rac/ROP small GTPases, NADPH oxidases, Pti-like kinases, harpin-induced 1 domain-containing proteins, and calcium-dependent protein kinases (Table 1). These proteins may therefore be important for PM microdomain-mediated disease resistance.
Table 1. Candidate Defense-Related Proteins That Are More Abundant in the Wild Type Than in OsFAH1/2-KD1 DRMs.
| Accession No. | Protein | emPAI Score |
Ratio (WT/KD) | ||
|---|---|---|---|---|---|
| WT | KD | ||||
| Receptor | gi|13324792 | Putative receptor kinase | 1.72 | 0.73 | 2.36 |
| gi|29367569 | Putative receptor protein kinase | 0.77 | 0.18 | 4.28 | |
| gi|218199769 | Cysteine-rich receptor-like protein kinase 10-like | 0.27 | – | – | |
| gi|46805208 | Putative protein kinase Xa21 | 0.09 | – | – | |
| gi|38345533 | Putative leucine-rich repeat receptor-like protein kinase family | 0.13 | – | – | |
| gi|125559260 | Putative receptor-like protein kinase 4 | 0.05 | – | – | |
| gi|215769298 | Receptor-like protein kinase family | 0.05 | – | – | |
| gi|222623298 | Putative serine/threonine kinase | 0.10 | – | – | |
| gi|218200640 | LRR receptor-like serine/threonine-protein kinase | 0.19 | – | – | |
| gi|15128407 | Putative receptor-like protein kinase | 0.05 | – | – | |
| gi|22748334 | Putative leucine-rich repeat transmembrane protein kinase | 0.11 | – | – | |
| gi|53792169 | Putative atypical receptor-like kinase MARK | 0.04 | – | – | |
| LRR-containing protein | gi|77552838 | Leucine-rich repeat family protein, expressed | 1.28 | 0.42 | 3.05 |
| gi|108863916 | Leucine-rich repeat family protein, expressed | 1.15 | – | – | |
| gi|77551698 | Leucine-rich repeat family protein, expressed | 0.05 | – | – | |
| gi|77548313 | Leucine-rich repeat family protein, putative, expressed | 0.24 | – | – | |
| gi|108707660 | Leucine-rich repeat family protein, expressed | 0.19 | – | – | |
| Chaperone | gi|115456247 | Similar to non-cell-autonomous heat shock cognate protein 70 | 0.29 | – | – |
| gi|115486793 | Heat shock protein 70 | 0.29 | – | – | |
| gi|115452223 | Similar to HSP70 | 0.22 | – | – | |
| gi|115464027 | Similar to HSP70 precursor | 0.10 | – | – | |
| Small GTPase | gi|115464861 | Small GTP-binding protein (OsRac2) | 0.80 | 0.34 | 2.35 |
| gi|115448617 | Small GTP-binding protein (OsRac3) | 0.34 | – | – | |
| gi|215678674 | RAC-ROP-like G-protein (OsRac1) | 0.56 | – | – | |
| NADPH oxidase | gi|77556657 | Respiratory burst oxidase, putative, expressed (OsRbohH) | 1.25 | 0.04 | 31.3 |
| gi|108864453 | Respiratory burst oxidase protein D, putative, expressed (OsRbohI) | 0.19 | 0.07 | 2.71 | |
| Pti-like kinase | gi|115456539 | Similar to Pti1 kinase-like protein | 1.61 | 0.30 | 5.34 |
| gi|115441637 | Similar to Pti1 kinase-like protein | 0.42 | – | – | |
| Harpin-induced 1 protein | gi|115487442 | Harpin-induced 1 domain containing protein | 0.96 | – | – |
| gi|77552873 | Harpin-induced protein 1 containing protein, expressed | 0.76 | – | – | |
| CDPK | gi|115472353 | Calcium-dependent protein kinase, isoform 2 (CDPK2) | 0.44 | 0.13 | 3.39 |
| gi|115456103 | Similar to calcium-dependent protein kinase | 0.13 | 0.06 | 2.17 | |
| Others | gi|115489172 | Thaumatin, pathogenesis-related family protein | 0.34 | – | – |
| gi|115467902 | Similar to resistant protein candidate | 0.17 | 0.08 | 2.13 | |
| gi|311893433 | Resistant protein | 0.02 | – | – | |
Experiments were repeated twice, and the proteins present at a level in the wild type more than double the level in OsFAH1/2-KD1 in both experiments are shown. The exponentially modified protein abundance index (emPAI) score indicates the score in a second set of experiments. LRR, leucine-rich repeat; WT, wild type.
PM Microdomains Are Required for the Localization and Specific Movement of Os-Rac1 and Os-Rbohs in the PM
Our proteome analysis revealed that Os-Rac1 is among the defense-related proteins that are enriched in PM microdomains (Table 1). Os-Rac1 plays a central role in rice innate immunity by upregulating various downstream components such as Rbohs (Kawano and Shimamoto, 2013; Kawano et al., 2014a). We found that two Rbohs (Os-RbohH and Os-RbohI) were also much less abundant in DRMs of OsFAH1/2-KD1 than the wild type (Table 1), suggesting that PM microdomains regulate innate immunity through the Rac1-Rboh-mediated pathway in rice. In a previous study, we found that myc-OsRac1 overexpressed in rice suspension cells shifted to the DRM fraction from the detergent-soluble membrane (non-DRM) fraction of the PM after treatment with chitin elicitor, a major component of the fungal cell wall that functions as a MAMP (Fujiwara et al., 2009). We therefore assessed the importance of PM microdomains for the localization and dynamics of Os-Rac1 and Os-Rbohs in response to chitin.
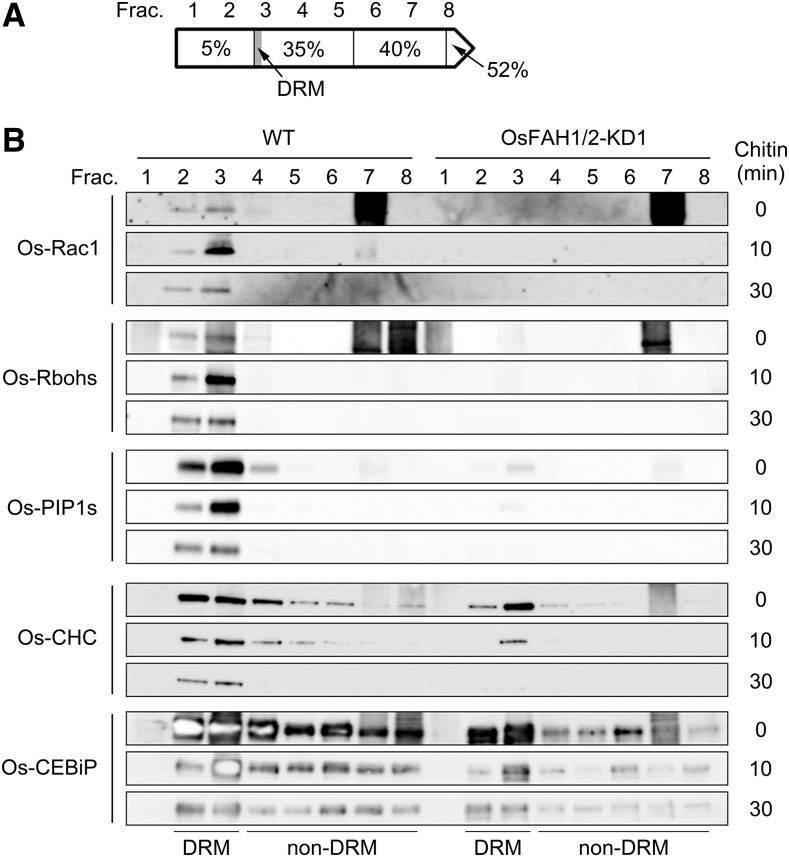
After the PM was extracted from wild-type and OsFAH1/2-KD1 suspension cells treated with chitin for 0, 10, or 30 min, DRM and non-DRM fractions were separated by OptiPrep density gradient centrifugation; DRMs existed in fractions 2 and 3 and non-DRMs in fractions 4 to 8 (Figure 5A). As shown in Figure 4B, endogenous Os-Rac1 appeared predominantly in the non-DRM fraction but also in the DRM fraction of wild-type cells before chitin treatment, as detected by α-OsRac1 antibody. Like overexpressed Os-Rac1, endogenous Os-Rac1 also switched dramatically to the DRM fraction of wild-type PMs after chitin treatment for 10 min and returned to normal levels in the DRM fraction after 30 min (Figure 5B). By contrast, no Os-Rac1 was detected in DRM fractions of the OsFAH1/2-KD1 line even after chitin treatment (Figure 5B). Interestingly, a similar result was obtained when Os-Rbohs were detected by an α-StRbohB antibody (Figure 5B). The accumulation of rice plasma membrane intrinsic protein 1s (Os-PIP1s), an aquaporin that localizes to PM microdomains (Hachez et al., 2013), was confirmed only in the DRM fraction of the wild type, whereas clathrin heavy chain (CHC), which is considered not to localize to PM microdomains (Li et al., 2012), was detected not only in the non-DRM fraction but also in the DRM fraction of both the wild type and OsFAH1/2-KD1 (Figure 5B). In addition, levels of both proteins gradually decreased during chitin treatment (Figure 5B). These results indicate that Os-Rac1 and Os-Rbohs are specific PM microdomain proteins that respond to chitin elicitor and that PM microdomains are necessary for their localization and dynamics in the PM. Moreover, Os-RACK1 and Os-HSP70, which interact with Os-Rac1 (Thao et al., 2007; Nakashima et al., 2008), and Os-flotillin, a PM microdomain protein (Li et al., 2012), were detected in only DRM fraction of the wild type (Supplemental Figure 10). On the other hand, Os-CEBiP, a chitin receptor, was present in most fractions of both the wild type and OsFAH1/2-KD1 and declined with chitin treatment (Figure 5B; Supplemental Figure 10).
Figure 5.
Dynamics of Os-Rac1 and Os-Rbohs in OsFAH1/2-KD1.
(A) Schematic image of OptiPrep gradient density centrifugation. Each gradient was separated into eight fractions from top (1) to bottom (8). Fractions 2 and 3 contain DRMs and fractions 4 to 8 contain the non-DRM fraction.
(B) Immunoblot analysis of Os-Rac1, Os-Rbohs, Os-CEBiP, Os-PIP1s, and Os-CHC after treatment with chitin for 0, 10, or 30 min.
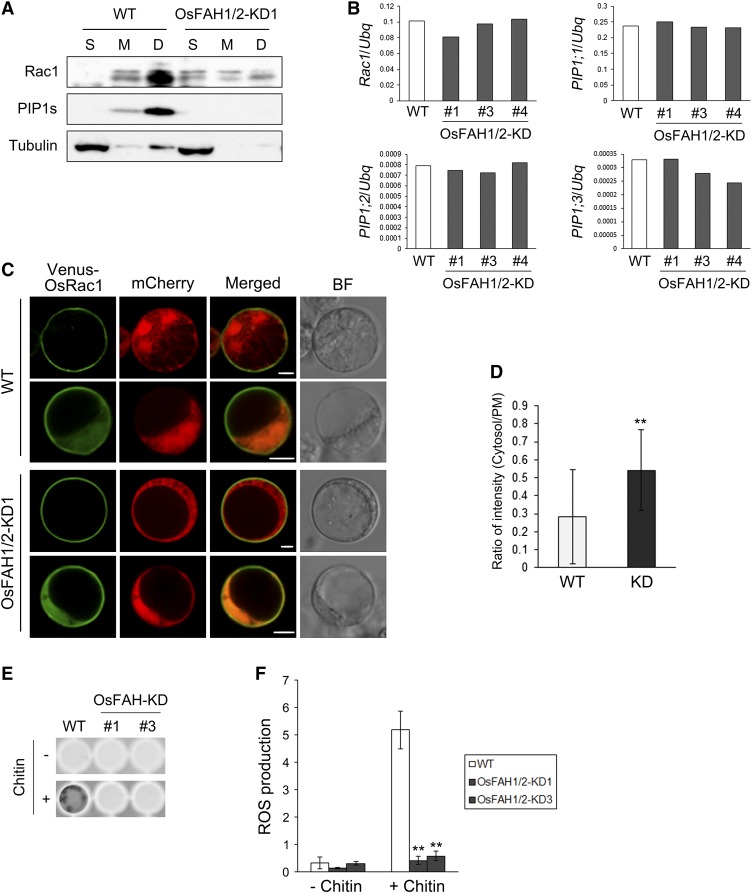
To explore the significance of Os-FAHs in the localization of Os-Rac1, we fractionated rice suspension cells into soluble, microsomal, and microsomal DRM-like fractions (Supplemental Figure 11) and examined the intracellular localization of Os-Rac1. As shown in Figure 6A, Os-Rac1 accumulated mainly in the microsomal DRM-like fraction of the wild type, and not in the soluble fraction. However, Os-Rac1 was detected in the soluble fraction of the OsFAH1/2-KD1 line and decreased in the microsomal DRM-like fraction. Similar to the result in Figure 5B, Os-PIP1s were most abundant in the microsomal DRM-like fraction of the wild type and were undetectable in any fractions of the OsFAH1/2-KD1 line (Figure 6A). Quantitative real-time PCR (qRT-PCR) showed that the expression of Os-Rac1 was normal in OsFAH1/2-KD1 (Figure 6B). Moreover, when YFP Venus-OsRac1 was expressed in protoplasts isolated from wild-type and the OsFAH1/2-KD suspension cells, the fluorescence patterns were divided into two types—only PM and PM, cytosol, and nucleus—as described by Chen et al. (2010a) (Figure 6C). However, the ratio of cytosol-localized to PM-localized Os-Rac1 in OsFAH1/2-KD1 was higher than in the wild type (Figure 6D). These data indicate that the membrane localization of Os-Rac1 is dependent on Os-FAHs.
Figure 6.
Intracellular Localization of Os-Rac1 and Production of ROS in the OsFAH1/2-KD.
(A) Immunoblot analysis of Rac1, PIP1s, and tubulin in wild-type and OsFAH1/2-KD1 suspension cells. S, M, and D indicate soluble, microsomal, and microsomal DRM-like fractions, respectively.
(B) qRT-PCR of Rac1, PIP1;1, PIP1;2, and PIP1;3 in wild-type and OsFAH1/2-KD1 suspension cells.
(C) Subcellular localization of Venus-OsRac1 in rice protoplasts extracted from the wild type and OsFAH1/2-KD1. The upper row for each cell type exemplifies localization mainly at the PM, and the lower row exemplifies localization mainly in the cytosol and nucleus. mCherry was used as a cytosolic and nuclear marker. BF, bright field. Bars = 5 μm.
(D) Ratio of the fluorescence intensity of the cytosol to that of the PM. Data are means ± sd (n ≥ 18). Asterisks indicate a significant difference compared with the wild type (Student’s t test; **P < 0.01).
(E) Example of ROS detection in rice suspension cells by L-012 reagent.
(F) Quantification of ROS production in OsFAH1/2-KD suspension cells. Intensity of each well was measured by ImageJ software (http://rsbweb.nih.gov/ij/). Data are means ± sd (n = 4). Asterisks indicate significant differences compared with the wild type (Student’s t test; **P < 0.01).
Os-FAHs Are Involved in ROS Production after Chitin Treatment
Os-Rac1 interacts with the N-terminal EF-hand motif of Os-RbohB, leading to an increase in ROS production (Wong et al., 2007; Oda et al., 2010; Kosami et al., 2014). To understand whether PM microdomains are necessary for ROS production mediated by the Rac1-Rboh pathway, we measured ROS production in OsFAH1/2-KD using the L-012 reagent. As shown in Figures 6E and 6F, wild-type suspension cells produced ROS in abundance after treatment with chitin elicitor, whereas ROS production in OsFAH1/2-KD cells was suppressed. This result indicates that Os-FAHs are crucial for ROS production in response to chitin treatment. Taken together with immunoblot analysis of the DRM fraction (Figure 5B), these data suggest that the localization of Os-Rac1 and Os-Rbohs to PM microdomains is essential for the production of ROS in response to chitin signaling.
Os-RbohB and Os-RbohH Are Major Defense-Related NADPH Oxidases Localized to PM Microdomains
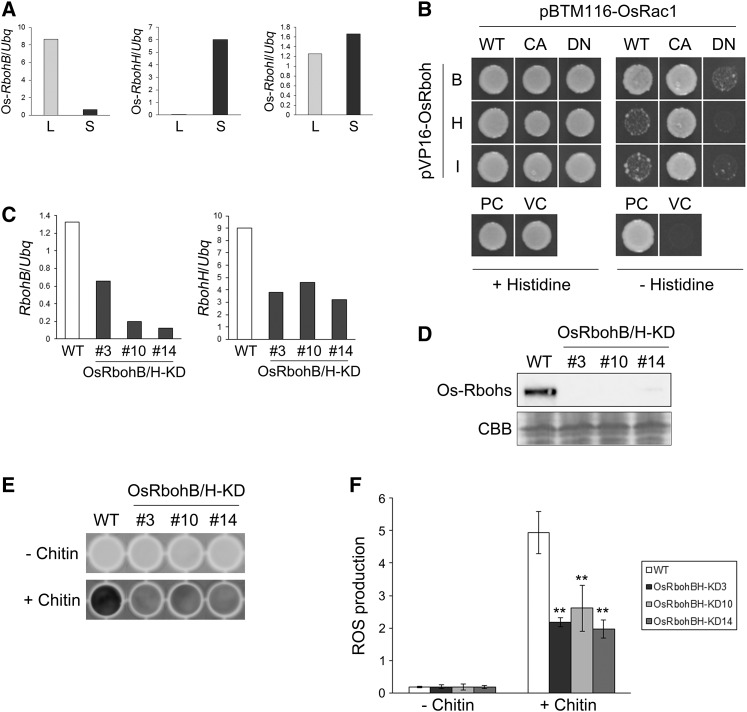
The rice genome encodes nine Rbohs (RbohA-RbohI), and our proteome analysis showed that Os-RbohH and Os-RbohI were present in PM microdomains of suspension cells (Table 1). Os-RbohH and Os-RbohI share a high degree of sequence identity with Os-RbohB and potato (Solanum tuberosum) St-RbohB (Supplemental Figure 12), which interact specifically with GTP-bound OsRac1 (Wong et al., 2007). Os-RbohH and Os-RbohI were also highly expressed in suspension cells, whereas the expression of Os-RbohB was higher in leaves, but lower in suspension cells (Figure 7A; Supplemental Figure 13A). These findings imply that Os-RbohH and Os-RbohI are PM microdomain-localizing NADPH oxidases activated by Os-Rac1 in rice suspension cells.
Figure 7.
Analysis of Os-RbohB and Os-RbohH.
(A) Expression of Os-RbohB, Os-RbohH, and Os-RbohI in rice leaves (L) and suspension cells (S) was analyzed by qRT-PCR.
(B) Interaction of Os-Rbohs with Os-Rac1 in the yeast two-hybrid system. Yeast cells containing bait and prey constructs were tested for binding on minimum medium without histidine. The images display the growth of yeast diluted at OD600 = 0.5 after 3 d of incubation at 30°C. Bait and prey vectors were pVP16 and pBTM116, respectively. CA, constitutively active; DN, dominant negative. The combination of pVP16-CA-OsRac1 and pBTM116-PAK CRIB was used as a positive control (PC), and the combination of empty vectors was used as a vector control (VC).
(C) Transcriptional analysis of RbohB and RbohH in OsRbohB/H-KD suspension cells by qRT-PCR.
(D) Immunoblot analysis of Os-Rbohs in DRMs extracted from wild-type and OsRbohB/H-KD suspension cells. An SDS-PAGE gel stained with Coomassie blue (CBB) was used to visualize protein loading.
(E) Example of ROS detection in OsRbohB/H-KD suspension cells by L-012 reagent.
(F) Quantification of ROS production in OsRbohB/H-KD suspension cells. Intensity of each well was measured by ImageJ software (http://rsbweb.nih.gov/ij/). Data are means ± sd (n = 4). Asterisks indicate significant differences compared with the wild type (Student’s t test; **P < 0.01).
To prove this, we first tested the interaction between Os-RbohH/I and Os-Rac1 by a yeast two-hybrid assay. As demonstrated by Wong et al. (2007), Os-RbohB interacted with constitutively active (CA)-OsRac1 but not with dominant negative (DN)-OsRac1 (Figure 7B), as did both Os-RbohH and Os-RbohI. By contrast, weak interactions between Os-RbohH/I and WT-OsRac1 were detected unlike Os-RbohB (Figure 7B; Supplemental Figure 13B). These results suggest that Os-RbohH and Os-RbohI interact with Os-Rac1 but that the intensity of the interaction is weaker than that between Os-RbohB and Os-Rac1.
Next, we investigated the effect of Os-RbohH on ROS production in the defense response in rice suspension cells because the amounts of protein and mRNA of Os-RbohH were much higher than those of Os-RbohI (Figure 7A, Table 1; Supplemental Figure 13A). We produced OsRbohH-knockdown (OsRbohH-KD) suspension cell lines using an RNAi system and ascertained that Os-RbohH expression decreased in these lines (Supplemental Figure 14A). However, ROS production in three OsRbohH-KD lines was equal to that in the wild type both with and without chitin treatment (Supplemental Figures 14B and 14C). To explain this unexpected result, we examined the expression of other Os-Rbohs and found that Os-RbohB was abnormally expressed in the OsRbohH-KD lines (Supplemental Figures 14D and 14E). We therefore also produced Os-RbohB and Os-RbohH double knockdown (OsRbohB/H-KD) suspension cell lines using RNAi and found that expression of both Os-RbohB and Os-RbohH decreased (Figure 7C). Immunoblot analysis confirmed that the OsRbohs were undetectable with an α-StRbohB antibody in DRMs of these OsRbohB/H-KD lines (Figure 7D). In addition, the OsRbohB/H-KD lines produced less ROS than the wild type after chitin treatment (Figures 7E and 7F). These findings indicate that Os-RbohB and Os-RbohH are the major chitin-responsive NADPH oxidases in rice PM microdomains.
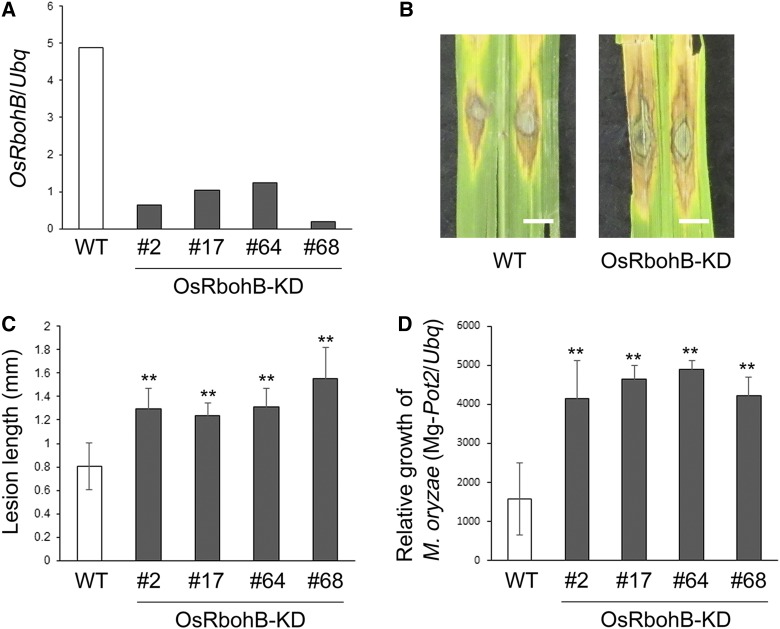
To test the relationship between Os-RbohB/H and rice innate immunity, we generated OsRbohB-knockdown plants (OsRbohB-KD; Figure 8A) because Os-RbohB was highly expressed in rice leaves and the expression of Os-RbohH was low (Figure 7A; Supplemental Figure 13A). When plants were infected with compatible rice blast fungus (race 007.0), lesion length in OsRbohB-KD was greater than in the wild type, with more extensive fungal growth (Figures 8B to 8D), suggesting that Os-RbohB is essential for resistance to rice blast fungus. In summary, we conclude that 2-hydroxy sphingolipid-rich PM microdomains play an essential role in chitin-triggered immunity by stimulating ROS production through the Rac1-RbohB/H pathway (Figure 9).
Figure 8.
Infection Assay for Rice Blast Fungus in OsRbohB-KD Plants.
(A) Transcriptional analysis of Os-RbohB in OsRbohB-KD plants by qRT-PCR.
(B) Infection of wild-type and OsRbohB-KD plants with a compatible strain of rice blast fungus (race007.0). Bars = 3 mm.
(C) Lesion length at 6 d postinfection in leaves of wild-type and OsRbohB-KD plants infected by the blast fungus. Data are means ± sd (n ≥ 30). Asterisks indicate significant differences compared with the wild type (Student’s t test; **P < 0.01).
(D) Relative growth of M. oryzae on susceptible rice cultivars. Data are means ± sd (n = 6). Asterisks indicate significant differences compared with the wild type (Student’s t test; **P < 0.01).
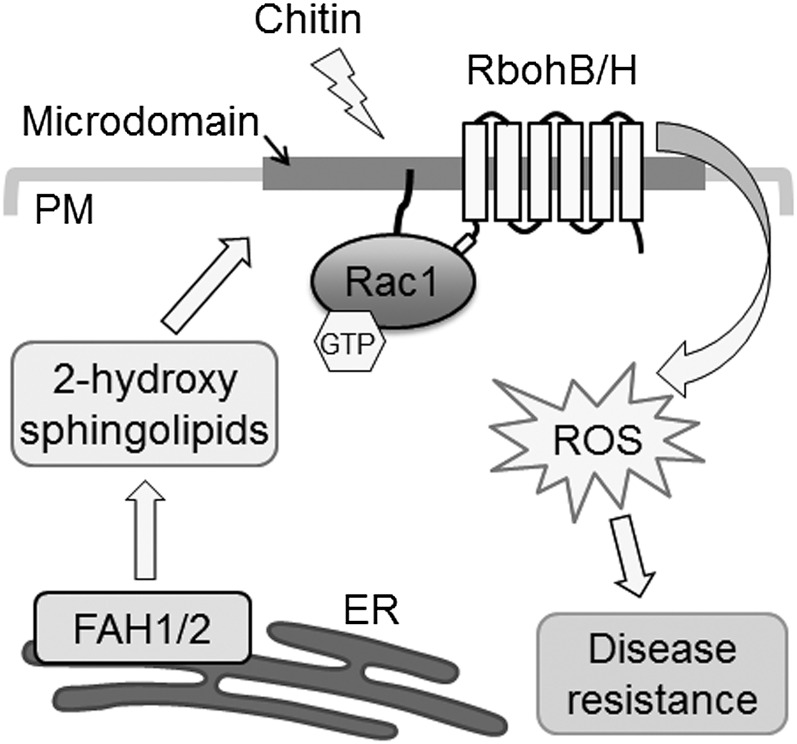
Figure 9.
Model of the Relationship among 2-Hydroxy Sphingolipids, PM Microdomains, and Innate Immunity in Rice.
Os-FAH1 and Os-FAH2, which are localized in the ER membrane, synthesize 2-hydroxy sphingolipids, which contribute to the organization of PM microdomains. After chitin treatment, Os-Rac1 and Os-RbohB/H assemble in microdomains, where Os-RbohB/H are activated by their interaction with GTP-bound Os-Rac1. This promotes the generation of ROS, leading to resistance to rice blast fungus infection.
DISCUSSION
Importance of PM Microdomain Organization by 2-Hydroxy Sphingolipids in Rice Cells
In artificial model membranes, sphingolipids and sterols display self-associative behavior, leading to their selective lateral segregation in the membrane or the microdomain concept (Lingwood and Simons, 2010). In plants, lipid analyses confirm the accumulation of sphingolipids, especially GlcCers and GIPCs, and sterols in PM microdomains (Lefebvre et al., 2007; Laloi et al., 2007; Cacas et al., 2016). In addition, depletion of sterols by treatment with agents such as MβCD and filipin has a severe effect not only on formation of PM microdomains, but also on microdomain protein composition and physiological functions including pollen tube tip growth and abscisic acid signaling (Roche et al., 2008; Kierszniowska et al., 2009; Liu et al., 2009; Demir et al., 2013). However, evidence of a relationship between sphingolipids and PM microdomain formation in plant cells is sparse. In this study, we revealed that 2-hydroxy sphingolipids are essential for the formation of PM microdomains in rice cells.
2-HFAs are major and specific fatty acid components of plant sphingolipids. For example, over 90% of complex sphingolipids in Arabidopsis possess 2-HFAs (Markham and Jaworski, 2007), and hGlcCers and hGIPCs are abundant in rice, which unlike Arabidopsis also contains nGIPCs. In this study, double knockdown of Os-FAH1 and Os-FAH2 led to a decrease in 2-hydroxy sphingolipids and an increase in non-hydroxy sphingolipids in cells. However, the levels of not only 2-hydroxy sphingolipids but also non-hydroxy sphingolipids and sterols decreased in DRMs of OsFAH1/2-KD cells. Because DRMs include sphingolipid- and sterol-enriched PM microdomains (Cacas et al., 2012), these results suggest that the decrease in 2-hydroxy sphingolipids by knockdown of Os-FAHs decreased the amount of DRMs, and especially PM microdomains. We also found that rice PIP1s, members of a family of PM microdomain proteins (Hachez et al., 2013), were not present in the DRM fraction of OsFAH1/2-KD, although Os-CHC, which does not localize in PM microdomains (Li et al., 2012), was detected, supporting the idea that PM microdomains would decrease in the DRM fraction of OsFAH1/2-KD. In addition, in vivo visualization of PM order using di-4-ANEPPDHQ demonstrated that the heterogeneity of the rice cell PM was lower in OsFAH1/2-KD1 than in the wild type. Furthermore, Cacas et al. (2016) recently confirmed the enrichment of hGIPCs in DRMs of tobacco (Nicotiana tabacum) cells. Therefore, hydroxyl groups in 2-hydroxy sphingolipids are likely to be important for the organization of PM microdomains in rice cells. Hydroxyl groups in sphingolipid fatty acids bind strongly to each other through hydrogen bonds (Pascher and Sundell, 1977; Löfgren and Pascher, 1977; Boggs et al., 1988), and hydroxyl groups are also largely present in LCBs of sphingolipids as dihydroxy (d18:0, d18:1, and d18:2) or trihydroxy LCBs (t18:0 and t18:1), and in sterols (Cacas et al., 2012). Thus, hydroxyl groups in 2-hydroxy sphingolipids may be required for the association of other sphingolipids and sterols, contributing to the rigidity of PM microdomains.
Our proteome analysis also demonstrated that cytoskeletal components such as tubulin and actin largely decreased in the DRM fraction of OsFAH1/2-KD (Table 1). Szymanski et al. (2015) recently demonstrated that actin filaments are related to the formation and dynamics of microdomains and that microtubules regulate the amounts of microdomain proteins by observation of microdomain marker proteins such as Remorin1.2 during deletion of actin filaments or microtubules. Therefore, the proper organization of PM microdomains would need not only lipids such as sphingolipids but also proteins, especially cytoskeleton. It will be interesting to examine the association between sphingolipids and the cytoskeleton in the formation of microdomains. In addition, cell wall could restrict the dynamics of PM proteins (Martiniere et al., 2012), suggesting that the cell wall is also involved in the organization of PM microdomains via PM proteins. Because we visualized PM microdomains of protoplasts in this study, the effect of the cell wall on the formation of PM microdomains remains unexplored. To address this, it will be necessary to observe microdomains in rice cells with a cell wall.
Observation of PM microdomain proteins by immunoelectron microscopy or stimulated emission-depletion microscopy has shown that the diameter of PM microdomains in plant cells is <100 nm (Raffaele et al., 2009; Li et al., 2012; Demir et al., 2013). In this study, PM microdomains themselves were observed as highly ordered puncta by the combination of di-4-ANEPPDHQ and TIRFM and were ∼250 nm in diameter. This may be because the high background in the surrounding membrane stained by di-4-ANEPPDHQ obscures the boundaries of puncta. Alternatively, actual PM microdomains may be larger than the clusters of PM microdomain proteins that they encompass. TIRFM is a very efficient microscopic technique that enables detailed observation of the PM and the simultaneous detection of two fluorescence emission wavelengths. However, improved microscopy or technology may be needed to observe PM microdomains more clearly using di-4-ANEPPDHQ.
Role of PM Microdomains in the Rac1-RbohB/H-Mediated MTI Pathway
Although PM microdomains are considered to play an important role in innate immunity in plants, their specific contribution has remained unclear. In this study, we demonstrated that susceptibility to compatible blast fungus infection was higher in OsFAH1/2-KD than in the wild type. Whereas PM microdomains, which are mainly formed by GlcCers and GIPCs in sphingolipids, decreased in OsFAH1/2-KD as described above, the hCer content also decreased, although the nCer content was not affected in whole cells. nCers have been reported to function as signaling molecules that promote programmed cell death, whereas hCers do not (Townley et al., 2005), implying that the decrease in hCers has little effect on the high sensitivity to rice blast fungus of OsFAH1/2-KD. In addition, OsFAH1/2-KD exhibited a dwarf phenotype; we used similarly sized leaves to the wild type during the infection assay to prevent an influence of the difference in growth. Therefore, the results of the infection assay of OsFAH1/2-KD suggest that PM microdomains play a key role in innate immunity in rice.
Focusing on Os-Rac1 and Os-Rbohs among the defense-related PM microdomain proteins identified by proteome analysis, we found that the decrease in PM microdomains diminished their localization to DRMs in response to chitin elicitor. In addition, the OsFAH1/2-KD lines displayed low ROS production. We further demonstrated that OsRbohB and OsRbohH are chitin-responsive NADPH oxidases that interact with Os-Rac1 and that Os-RbohB is essential for resistance to rice blast fungus. These findings indicate that PM microdomains are essential for chitin-induced MTI through ROS signaling mediated by Os-Rac1 and Os-RbohB/H.
Os-Rac1 plays a critical role in defense responses induced by various MAMPs, including chitin elicitor (Kawano and Shimamoto, 2013; Kawano et al., 2014a). After chitin is recognized by Os-CEBiP, which localizes to the extracellular side of the PM, Os-Rac1 is converted from the GDP to the GTP form by the guanine nucleotide exchange factor Os-RacGEF1, which is phosphorylated by Os-CERK1, a coreceptor of chitin (Kaku et al., 2006; Shimizu et al., 2010; Akamatsu et al., 2013). In this study, we provided evidence that endogenous Os-Rac1 is transiently localized to PM microdomains at an early stage of the chitin response. Os-Rac1 belongs to the type II Rac/ROP subgroup, whose members are attached to the PM by palmitoylation of a conserved GC-CG box (Lavy et al., 2002; Levental et al., 2010). Palmitoylation plays a key role in targeting proteins to PM microdomains (Levental et al., 2010). Arabidopsis ROP6, a type I ROP possessing a CaaL box motif, is primarily geranylgeranylated and, when activated, accumulates in DRMs with transient palmitoylation (Sorek et al., 2007, 2010). In addition, mammalian Rac1 is palmitoylated at C178, a modification that is essential for targeting to rafts and actin cytoskeleton remodeling (Navarro-Lérida et al., 2012), and present in not only rafts but also non-rafts, where Rac1 is predominantly inactivated (Moissoglu et al., 2014). Activation of Os-Rac1 occurs within 3 min after chitin treatment (Akamatsu et al., 2013), suggesting that GTP-bound Os-Rac1 is localized to PM microdomains, since Os-Rac1 had accumulated in the DRM fraction by 10 min of chitin treatment. We therefore propose that the localization of Os-Rac1 to PM microdomains, possibly through palmitoylation, is necessary for its function, similar to other small GTPases. By contrast, Os-Rac1 was accumulated in the “DRM-like” fraction in Figure 6A, which is crudely extracted from microsomes as shown in Supplemental Figure 11, without chitin treatment. Because the DRM fraction in Figure 5B is carefully extracted from the PM, the behavior of Os-Rac1 on PM microdomains can be interpreted in Figure 5B more correctly than in Figure 6A.
We also demonstrated that Os-RbohB and Os-RbohH display similar dynamics to Os-Rac1 in PM microdomains in response to chitin. OsRbohB acts downstream of Os-Rac1 through a direct interaction of its N-terminal EF-hand motif with GTP-bound Os-Rac1 (Wong et al., 2007; Oda et al., 2010; Kosami et al., 2014), and, as shown in this study, Os-RbohH is also involved in Rac1-mediated immunity. Proteome analyses have identified Rbohs in DRMs of tobacco, Arabidopsis and rice (Morel et al., 2006; Kierszniowska et al., 2009; Fujiwara et al., 2009), and ROS production by Rbohs in pollen tube tips is dependent on PM microdomains in Picea meyeri (Liu et al., 2009). Recent reports also showed that the dynamics of AtRbohD are regulated by PM microdomains (Hao et al., 2014). At-RbohD has been shown to play an important role in innate immunity in Arabidopsis (Kadota et al., 2015) and belongs to same clade with Os-RbohB and Os-RbohH (Supplemental Figure 11A). Taken together with the data that OsRbohB contributes to resistance to rice blast fungus infection, simultaneous localization of Os-Rac1 and Os-RbohB/H to PM microdomains in response to chitin is likely to be necessary for the activation of Os-RbohB/H by activated Os-Rac1. Bioimaging analysis to observe the dynamics and colocalization of Os-Rac1 and Os-RbohB/H is needed. On the other hand, the amount of Os-Rbohs in non-DRM fraction of OsFAH1/2-KD without chitin treatment was lower than in the wild type, suggesting that the decrease in 2-hydroxy sphingolipids may also affect the secretion pathway of PM proteins, in which sphingolipids are involved (Luttgeharm et al., 2016). Future work should examine the relationship among 2-hydroxy sphingolipids, secretion of Os-Rbohs and ROS production.
We have previously proposed that rice Rac1 participates in a large protein network, termed the defensome, including HSP70, HSP90, the cochaperone Hop/Sti1a, the scaffold protein RACK1, and the lignin biosynthesis enzyme CCR1 (Kawasaki et al., 2006; Thao et al., 2007; Nakashima et al., 2008; Chen et al., 2010b). Fujiwara et al. (2009) demonstrated that RACK1 shifts to the DRM fraction in response to chitin treatment, similar to Rac1. In this study, we also demonstrate that HSP70 as well as RACK1 localize to PM microdomains. Therefore, a number of other defensome proteins may localize to PM microdomains with Rac1. It will be important to establish whether and how PM microdomains are related to the formation of the defensome network. It will also be interesting to examine the correlation between PM microdomains and the ETI pathway because rice Rac1 is regulated by the ETI-associated protein Pit, which is localized to the PM (Kawano et al., 2010, 2014b). We cannot rule out the possibility that PM microdomains could pleiotropically affect plant innate immunity since our proteome analysis detected a large number of defense-related proteins in addition to Rac1 and Rbohs. Therefore, it will be necessary to investigate the role of other microdomain proteins in rice innate immunity.
In this report, we established OsFAH1/2-KD lines in which PM microdomains are constitutively decreased and were able to reveal the importance of PM microdomains in chitin-induced immunity because these lines could be used in physiological experiments. In addition, we showed that PM microdomains may be important for growth. Further studies using OsFAH1/2-KD lines will deepen our understanding of the significance of PM microdomains in plants.
METHODS
Plant Materials
Rice (Oryza sativa japonica Kinmaze) was used as the wild type. To generate constructs for the RNAi system, cDNA fragments of the 3′ untranslated region of Os-FAH1/2 or Os-RbohB/H (primer sets are described in Supplemental Table 1) were introduced into the pANDA vector (Miki and Shimamoto, 2004). Agrobacterium tumefaciens-mediated transformation of rice calli was performed according to a published method (Hiei et al., 1994). Plants were regenerated from transformed calli selected by hygromycin resistance. Rice plants were cultivated at 27°C under short-day conditions (10 h light/14 h dark) with sodium lamps (200 μmol m−2 s−1).
Intracellular Localization
Constructs encoding TagRFP, enhanced GFP, or Venus fused to either the C or N terminus of Os-FAH1, Os-FAH2, and Os-Rac1 (primer sets are described in Supplemental Table 1) were prepared using the Gateway system (Invitrogen). OsFAH1-TagRFP, OsFAH2-TagRFP, OsFAH2-EGFP, and Venus-OsRac1 expression was driven by the CaMV 35S promoter. Protoplast isolation from rice suspension cultures and protoplast transformation were performed according to Chen et al. (2010a). After incubation for 12 to 18 h at 30°C, the protoplasts were examined with a Leica TCS-SP5 microscope. A 488- and 514-nm argon laser were used for the excitation of GFP and YFP, respectively. A 543-nm H2/Ne laser was also used for RFP. Emission signals were detected using a 500/15-nm filter for GFP, a 515/30-nm filter for YFP, and a 590/70-nm filter for RFP.
Preparation of Sphingolipids and Sterols
Total lipids were extracted from rice cells (300 mg fresh weight) and treated with methylamine to remove glycerolipids, as described by Markham et al. (2006). An internal standard mixture was added during extraction, consisting of d18:1-c12:0 Cer (0.1 nmol for nCer), d18:1-h12:0 Cer (0.1 nmol for hCer), d18:1-c12:0 GlcCer (1 nmol for GlcCer), d17:1-c12:0 sphingosyl phosphatidylethanolamine (2 nmol for GIPC), β-cholestanol (50 nmol for free sterol [FS]), and cholestanol glucoside (15 nmol for steryl glucoside [SG]). Standard sphingolipids were purchased form Avanti Polar Lipids, and β-cholestanol was obtained from Sigma-Aldrich. Glucoside derivatives of β-cholestanol and other sterols were synthesized as described previously (Iga et al., 2005; Wewer et al., 2011). Alkali-treated lipid extract was dried, resuspended in 1 mL tetrahydrofuran (THF)/methanol/water (2:1:2, v/v/v) containing 0.1% formic acid, and used for LC-MS/MS analyses of sphingolipids and SG. FS was analyzed by LC-MS/MS after picolinyl esterification according to Honda et al. (2008).
LC-MS/MS Analysis of Sphingolipids and Sterols
Sphingolipids and sterols were quantified using an LCMS-8030 system (Shimadzu). Analytical conditions described for Arabidopsis thaliana by Markham and Jaworski (2007) and Nagano et al. (2014) were optimized for rice sphingolipid species. Chromatographic separation was performed using an XR-ODSII column (Shimadzu; 2.2 μm, 2.0 mm ID, 75 mm) held at 40°C with binary elution gradients consisting of THF/methanol/10 mM ammonium formate (3:2:5) containing 0.1% (v/v) formic acid as solvent A and THF/methanol/10 mM ammonium formate (7:2:1) containing 0.1% (v/v) formic acid as solvent B at a flow rate of 200 μL/min. Each lipid class (Cer, GlcCer, GIPC, SG, and FS) was analyzed separately using solvent concentration gradients, applied linearly over 15 min, as follows: 30 to 100% B for Cer, GlcCer, FS, and SG; 10 to 70% B for GIPC. After elution, the column was washed with 100% solvent B for 1 min and reequilibrated with the appropriate starting solvent for 3 min before the next run.
Lipid species were detected by multiple-reaction monitoring of the transitions of precursor ions [M+H]+ to main product ions. Major components of rice sphingolipids containing fatty acids with C20, C22, and C24 carbon length (Watanabe and Imai, 2011) were targeted. To compensate for differences in the MS responses of endogenous sphingolipid species and the internal standards added to samples, sphingolipid classes were fractionated from rice and MS response factors of each internal standard to endogenous species were determined on the basis of their LCB content according to Markham and Jaworski (2007). GIPC content is shown as the total of four subclasses with different sugar moieties (Supplemental Data Set 4). MS response factors for FSs and glucosides were determined using commercially available standards. The m/z values of multiple-reaction monitoring transitions and collision energy were optimized for each compound. Detailed MS/MS parameters for targeted species are listed in Supplemental Data Set 4. The other general conditions were as follows: capillary voltage, 4.5 kV; desolvation gas flow, 10 L/min; nebulizer gas flow, 0.2 L/min; conversion diode voltage, 6 kV; source temperature, 300°C; and collision gas flow, 230 kPa.
Staining with Di-4-ANEPPDHQ and Imaging of Membrane Order
Protoplasts were treated at 10 min with 10 μM di-4-ANEPPDHQ (Molecular Probes) dissolved in a medium at 10 min and excited with a 488-nm argon laser. For observation by CLSM (Leica TCS-SP5), Lo and Ld images were obtained by measuring fluorescence at 500 to 550 nm and 650 to 750 nm, respectively, in a modification of the methods described by Owen et al. (2011). For observation by TIRFM (Olympus TIRFM), the fluorescence emission spectra were separated with a 565LP dichroic mirror and filtered through either a 500/550 (Lo) or a 641.5/708.5 (Ld) filter with a Dual View filter system (Olympus). A calibration of di-4-ANEPPDHQ was performed according to the procedure described by Owen et al. (2011).
Extraction of PM and DRM
PM was purified from rice suspension cells using the two-phase partition method (Uemura et al., 1995). DRMs were extracted from the PM fraction in a modification of the methods described by Fujiwara et al. (2009). Briefly, the PM fractions were treated with Triton X-100 to a detergent:PM protein ratio of 15:1, and the mixture was shaken for 30 min at 4°C. The sample was diluted with OptiPrep solution to a final concentration of 52% OptiPrep (w/w), overlaid with 40, 35, and 5% OptiPrep in TED buffer (50 mM Tris-HCl, pH 7.5, 3 mM EDTA, and 1 mM DTT; w/w), and centrifuged for 16 h at 150,000g at 4°C in a Swi40Ti rotor (Beckman). DRM fractions were recovered above the 5 and 35% interface (Figure 5A), diluted 10 times with TED buffer and centrifuged for 2 h at 200,000g. The final pellets were suspended in TED buffer. Non-DRM fractions were recovered from 35, 40, and 52% OptiPrep fractions (fractions 4 to 8 in Figure 5A).
Peptide Preparation for MS/MS Analysis
DRMs were separated using a ready-made 12.5% (w/v) SDS-polyacrylamide gel (DRC) and stained using Flamingo (Bio-Rad). The gel lane was cut into four slices of equal length. Each gel band was washed twice with HPLC-grade water containing 60% (v/v) acetonitrile (Kanto Chemical)/50 mM ammonium bicarbonate, and incubated successively in 10 mM DTT/50 mM ammonium bicarbonate for 45 min at 56°C and 55 mM iodoacetamide/50 mM ammonium bicarbonate for 30 min at room temperature. The incubated gel slices were washed twice with HPLC-grade water containing 60% (v/v) acetonitrile/50 mM ammonium bicarbonate and dried in a vacuum concentrator. The dried slices were next treated with 2 μL of 10 ng/μL trypsin (MS grade gold; Promega)/50 mM ammonium bicarbonate and incubated at 37°C for 16 h. The digested peptides were recovered to a new tube. The gel was then treated twice with 20 μL of 0.2% (v/v) formic acid (Wako)/50% (v/v) acetonitrile, and the three peptide extracts were pooled. The extracts were dried in a vacuum concentrator and dissolved in 0.1% (v/v) formic acid/5% (v/v) acetonitrile. The solution was filtered with an Ultrafree-MS Centrifugal Filter (PVDF 0.45 μm; Millipore) to avoid contamination with gel pieces.
LC-MS/MS Analysis and Database Searching
LC-MS/MS analysis was performed using an HTC-PAL/Paradigm MS4 system coupled to an LTQ Orbitrap XL (Thermo Scientific) mass spectrometer. Trypsin-digested peptides were loaded on the L-column (100 μm internal diameter, 15 cm; CERI) using a Paradigm MS4 HPLC pump (Michrome BioResources) and an HTC-PAL autosampler (CTC Analytics). Buffers were 0.1% (v/v) acetic acid and 2% (v/v) acetonitrile in water (Solvent A) and 0.1% (v/v) acetic acid and 90% (v/v) acetonitrile in water (Solvent B). A linear gradient from 5 to 45% buffer B, of 26 min duration, was applied, and peptides eluted from the L-column were introduced directly into an LTQ Orbitrap XL mass spectrometer with a flow rate of 500 nL/min and a spray voltage of 2.0 kV. All events for MS scan were controlled and acquired by Xcalibur software version 2.0.7 (Thermo Scientific). The range of MS scan was m/z 400 to 1500 and the top three peaks were subjected to MS/MS analysis. Obtained spectra were compared with a protein database (NCBI, Taxonomy; Oryza sativa) using the MASCOT server (version 2.4). The mascot search parameters were as follows: peptide tolerance at 10 ppm, MS/MS tolerance at ±0.5 D, peptide charge of 2+ or 3+, trypsin as enzyme allowing up to one missed cleavage, carbamidomethylation on cysteine as a fixed modification, and oxidation on methionine as a variable modification.
Infection of Rice Plants with Rice Blast Fungus
Growth conditions for the blast fungus (Magnaporthe oryzae; strain 2403-1, race007.0) and methods for punch inoculation of leaf blades have been described previously (Thao et al., 2007). To measure M. oryzae growth, DNA-based real-time PCR was performed according to the method described by Kawano et al. (2010).
SA Analysis
SA was prepared using MTBE/methanol/water and quantified by LC-MS/MS according to Ternes et al. (2011).
Chitin Treatment
Cultured rice cells were treated with 2 μg/mL chitin (hexa-N-acetylchitohexaose; IsoSep AB) and harvested at the indicated times after treatment.
Antibodies
α-OsRac1, α-RACK1, and α-flotillin antibody were described previously (Lieberherr et al., 2005; Nakashima et al., 2008; Ishikawa et al., 2015). α-StRbohB, α-OsCEBiP, and α-CHC antibodies were kindly provided by Hirofumi Yoshioka (Nagoya University), Naoto Shibuya (Meiji University), and Masaru Fujimoto (University of Tokyo), respectively. α-OsPIP1s, α-tubulin, and α-HSP70 antibody were purchased from Cosmo Bio, Calbiochem, and Funakoshi, respectively.
Cell Fractionation
Rice cells in suspension cultures were harvested 4 d after subculturing and homogenized in homogenizing medium (50 mM Tris-HCl, pH 7.5, 2 mM EDTA, 150 mM NaCl, 5 mM MgCl2, 10% glycerol, and protease inhibitor cocktail [Roche]). After centrifugation at 8000g at 4°C for 20 min, the supernatant was ultracentrifuged at 100,000g for 1 h at 4°C and separated into supernatant and pellet. This supernatant was defined as the soluble fraction. The pellet was further treated with 1% Triton X-100, incubated on ice for 30 min, and centrifuged at 20,000g for 20 min at 4°C. The pellet was recovered as the DRM-like fraction and the supernatant as the microsomal fraction.
ROS Measurement
Rice suspension cells were subcultured for 4 d in fresh medium and placed into white 96-well plates (Greiner Bio-one). L-012 (Wako Chemicals), dissolved in culture medium to 500 μM, was added to each well, and after a 2-h incubation the chemiluminescence was detected by LAS-4000 mini luminescent image analyzer (Fujifilm). The intensity of each well was measured by ImageJ (http://rsbweb.nih.gov/ij/), and the value was calculated using the following formula: (intensity of each well – intensity of background)/weight of suspension cells in each well.
Yeast Two-Hybrid Assay
For bait constructs, WT-, CA-, and DN-OsRac1 were subcloned into the pBTM116 vector using the Gateway system (Invitrogen) as described by Kawasaki et al. (2006). For prey constructs, sequences encoding the N-terminal regions of Os-Rbohs were inserted into the pVP16 vector using the Gateway system (Invitrogen). Combinations of bait and prey vectors were introduced into Saccharomyces cerevisiae L40 cells. The bait/prey interaction was analyzed based on the requirement of His for yeast growth, as described by Wong et al. (2007).
qRT-PCR
Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen). cDNA was synthesized from 1 μg total RNA using SuperScript II reverse transcriptase (Invitrogen). qPCR reactions were performed with 7300 real-time PCR system (Applied Biosystems) and the primer sets shown in Supplemental Table 1.
Phylogenetic Analysis
Predicted amino acid sequences were aligned using the MUSCLE in the MEGA program (Kumar et al., 2016). Based on these alignments (Supplemental Data Set 5), a phylogenetic tree was generated using the Maximum Likelihood method with the MEGA program, and the reliability of the trees was tested by bootstrap with 1000 resamplings.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: Os-FAH1, Os12g0628400; Os-FAH2, Os03g0780800; Os-Rac1, Os01g0229400; Os-RbohA, NM_191558; Os-RbohB, Os01g0360200; Os-RbohC, AK120905; Os-RbohD, AK072353; Os-RbohE, AK100241; Os-RbohF, XM_482730; Os-RbohG, AK120739; Os-RbohH, Os12g0541300; Os-RbohI, Os11g0537400; Os-PIP1;1, Os02g0666200; Os-PIP1;2, Os04g0559700; Os-PIP1;3, Os02g0823100; St-RbohA, AB050660; and St-RbohB, AB050661.
Supplemental Data
Supplemental Figure 1. Examples of sphingolipids in plants.
Supplemental Figure 2. Subcellular localization of Os-FAH1 and Os-FAH2 in rice protoplasts.
Supplemental Figure 3. Establishment of OsFAH1/2-KD lines using an RNAi system.
Supplemental Figure 4. GIPC content in whole cells of OsFAH1/2-KD lines.
Supplemental Figure 5. Fatty acid content of sphingolipids in OsFAH1/2-KD lines.
Supplemental Figure 6. GIPC content in DRM of OsFAH1/2-KD1.
Supplemental Figure 7. Sphingolipid content in PM of OsFAH1/2-KD lines.
Supplemental Figure 8. GIPC content in PM of OsFAH1/2-KD1.
Supplemental Figure 9. SA content in OsFAH1/2-KD suspension cells.
Supplemental Figure 10. Protein analysis of PM fractions.
Supplemental Figure 11. Flow chart of fractionation of rice suspension cells.
Supplemental Figure 12. Phylogenetic relationships and comparison of Rboh proteins.
Supplemental Figure 13. Analysis of Os-Rbohs.
Supplemental Figure 14. Analysis of OsRbohH-KD lines.
Supplemental Data Set 1. Identification of DRM-associated proteins in the wild type.
Supplemental Data Set 2. Identification of DRM-associated proteins in OsFAH1/2-KD1.
Supplemental Data Set 3. Identification of DRM-associated proteins for which wild type > KD.
Supplemental Data Set 4. Parameters for LC-MS/MS analysis of rice sphingolipids and sterols.
Supplemental Data Set 5. Text file of the alignment used for the phylogenetic analysis shown in Supplemental Figure 12A.
Supplemental Table 1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Hirofumi Yoshioka (Nagoya University, Japan) for α-StRbohB antibody, Naoto Shibuya (Meiji University, Japan) for α-OsCEBiP antibody, Noriko Inada (Nara Institute of Science and Technology, Japan) for technical advice about TIRFM, and Masaru Fujimoto for α-CHC antibody and technical advice about TIRFM. We also thank Yuko Tamaki and Ms. Junko Naritomi (Nara Institute of Science and Technology) for rice transformation and Kaori Tashiro (Saitama University) for technical support. This research was supported by a Grant-in-Aid for JSPS Fellows Grant 238349 to M.N.; JSPS KAKENHI Grants 19108005 to K.S., 26292190 to M.K.-Y., 26450055 and 26113715 to Y.K., and 26850232 to M.N.; MAFF Genomics for Agricultural Innovation Grant PMI-0007 to K.S.; and Takeda Science Foundation, PSC, NSFC, and CAS Hundred Talents Program to Y.K.
AUTHOR CONTRIBUTIONS
M.N. and K.S. planned the experiments. M.N., T.I., M.F., and Y.F. carried out the experiments. M.N., Y.K. and M.K-Y. wrote the manuscript.
Glossary
- MAMP
microbe-associated molecular pattern
- MTI
MAMP-triggered immunity
- ROS
reactive oxygen species
- ETI
effector-triggered immunity
- PM
plasma membrane
- LCB
long-chain base
- GlcCer
glucosylceramide
- GIPC
glycosylinositolphosphoceramide
- 2-HFA
2-hydroxy fatty acid
- ER
endoplasmic reticulum
- NFA
non-hydroxy fatty acid
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- DRM
detergent-resistant membrane
- TIRFM
total internal reflection fluorescence microscopy
- CLSM
confocal laser scanning microscopy
- SA
salicylic acid
- FS
free sterol
- SG
steryl glucoside
- THF
tetrahydrofuran
References
- Akamatsu A., Wong H.L., Fujiwara M., Okuda J., Nishide K., Uno K., Imai K., Umemura K., Kawasaki T., Kawano Y., Shimamoto K. (2013). An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe 13: 465–476. [DOI] [PubMed] [Google Scholar]
- Antolín-Llovera M., Ried M.K., Binder A., Parniske M. (2012). Receptor kinase signaling pathways in plant-microbe interactions. Annu. Rev. Phytopathol. 50: 451–473. [DOI] [PubMed] [Google Scholar]
- Boggs J.M., Rangaraj G., Koshy K.M. (1988). Photolabeling of myelin basic protein in lipid vesicles with the hydrophobic reagent 3-(trifluoromethyl)-3-(m-[125I]iodophenyl)diazirine. Biochim. Biophys. Acta 937: 1–9. [DOI] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Cacas J.L., et al. (2016). Revisiting plant plasma membrane lipids in tobacco: A focus on sphingolipids. Plant Physiol. 170: 367–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacas J.L., Furt F., Le Guédard M., Schmitter J.M., Buré C., Gerbeau-Pissot P., Moreau P., Bessoule J.J., Simon-Plas F., Mongrand S. (2012). Lipids of plant membrane rafts. Prog. Lipid Res. 51: 272–299. [DOI] [PubMed] [Google Scholar]
- Chen L., Shiotani K., Togashi T., Miki D., Aoyama M., Wong H.L., Kawasaki T., Shimamoto K. (2010a). Analysis of the Rac/Rop small GTPase family in rice: expression, subcellular localization and role in disease resistance. Plant Cell Physiol. 51: 585–595. [DOI] [PubMed] [Google Scholar]
- Chen L., Hamada S., Fujiwara M., Zhu T., Thao N.P., Wong H.L., Krishna P., Ueda T., Kaku H., Shibuya N., Kawasaki T., Shimamoto K. (2010b). The Hop/Sti1-Hsp90 chaperone complex facilitates the maturation and transport of a PAMP receptor in rice innate immunity. Cell Host Microbe 7: 185–196. [DOI] [PubMed] [Google Scholar]
- Cui H., Tsuda K., Parker J.E. (2015). Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66: 487–511. [DOI] [PubMed] [Google Scholar]
- Demir F., Horntrich C., Blachutzik J.O., Scherzer S., Reinders Y., Kierszniowska S., Schulze W.X., Harms G.S., Hedrich R., Geiger D., Kreuzer I. (2013). Arabidopsis nanodomain-delimited ABA signaling pathway regulates the anion channel SLAH3. Proc. Natl. Acad. Sci. USA 110: 8296–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Rathjen J.P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11: 539–548. [DOI] [PubMed] [Google Scholar]
- Fujimoto M., Arimura S., Nakazono M., Tsutsumi N. (2007). Imaging of plant dynamin-related proteins and clathrin around the plasma membrane by variable incidence angle fluorescence microscopy. Plant Biotechnol. 24: 449–455. [Google Scholar]
- Fujiwara M., Hamada S., Hiratsuka M., Fukao Y., Kawasaki T., Shimamoto K. (2009). Proteome analysis of detergent-resistant membranes (DRMs) associated with OsRac1-mediated innate immunity in rice. Plant Cell Physiol. 50: 1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Chung E.H., Eitas T.K., Dangl J.L. (2011). Plant intracellular innate immune receptor Resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc. Natl. Acad. Sci. USA 108: 7619–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Zhou D., Pryse K.M., Okunade A.L., Su X. (2010). Fatty acid 2-hydroxylase mediates diffusional mobility of Raft-associated lipids, GLUT4 level, and lipogenesis in 3T3-L1 adipocytes. J. Biol. Chem. 285: 25438–25447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachez C., Besserer A., Chevalier A.S., Chaumont F. (2013). Insights into plant plasma membrane aquaporin trafficking. Trends Plant Sci. 18: 344–352. [DOI] [PubMed] [Google Scholar]
- Hao H., Fan L., Chen T., Li R., Li X., He Q., Botella M.A., Lin J. (2014). Clathrin and membrane microdomains cooperatively regulate RbohD dynamics and activity in Arabidopsis. Plant Cell 26: 1729–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282. [DOI] [PubMed] [Google Scholar]
- Honda A., Yamashita K., Miyazaki H., Shirai M., Ikegami T., Xu G., Numazawa M., Hara T., Matsuzaki Y. (2008). Highly sensitive analysis of sterol profiles in human serum by LC-ESI-MS/MS. J. Lipid Res. 49: 2063–2073. [DOI] [PubMed] [Google Scholar]
- Iga D.P., Iga S., Schmidt R.R., Buzas M.C. (2005). Chemical synthesis of cholesteryl β-d-galactofuranoside and -pyranoside. Carbohydr. Res. 340: 2052–2054. [DOI] [PubMed] [Google Scholar]
- Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M. (2005). Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4: 1265–1272. [DOI] [PubMed] [Google Scholar]
- Ishikawa T., Aki T., Yanagisawa S., Uchimiya H., Kawai-Yamada M. (2015). Overexpression of BAX INHIBITOR-1 links plasma membrane microdomain proteins to stress. Plant Physiol. 169: 1333–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y., Shirasu K., Zipfel C. (2015). Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 56: 1472–1480. [DOI] [PubMed] [Google Scholar]
- Kaku H., Nishizawa Y., Ishii-Minami N., Akimoto-Tomiyama C., Dohmae N., Takio K., Minami E., Shibuya N. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 103: 11086–11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y., Akamatsu A., Hayashi K., Housen Y., Okuda J., Yao A., Nakashima A., Takahashi H., Yoshida H., Wong H.L., Kawasaki T., Shimamoto K. (2010). Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Microbe 7: 362–375. [DOI] [PubMed] [Google Scholar]
- Kawano Y., Shimamoto K. (2013). Early signaling network in rice PRR-mediated and R-mediated immunity. Curr. Opin. Plant Biol. 16: 496–504. [DOI] [PubMed] [Google Scholar]
- Kawano Y., Kaneko-Kawano T., Shimamoto K. (2014a). Rho family GTPase-dependent immunity in plants and animals. Front. Plant Sci. 5: 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y., Fujiwara T., Yao A., Housen Y., Hayashi K., Shimamoto K. (2014b). Palmitoylation-dependent membrane localization of the rice resistance protein pit is critical for the activation of the small GTPase OsRac1. J. Biol. Chem. 289: 19079–19088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T., Koita H., Nakatsubo T., Hasegawa K., Wakabayashi K., Takahashi H., Umemura K., Umezawa T., Shimamoto K. (2006). Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc. Natl. Acad. Sci. USA 103: 230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath N.F., Kierszniowska S., Lorek J., Bourdais G., Kessler S.A., Shimosato-Asano H., Grossniklaus U., Schulze W.X., Robatzek S., Panstruga R. (2010). PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J. Biol. Chem. 285: 39140–39149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszniowska S., Seiwert B., Schulze W.X. (2009). Definition of Arabidopsis sterol-rich membrane microdomains by differential treatment with methyl-β-cyclodextrin and quantitative proteomics. Mol. Cell. Proteomics 8: 612–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König S., Feussner K., Schwarz M., Kaever A., Iven T., Landesfeind M., Ternes P., Karlovsky P., Lipka V., Feussner I. (2012). Arabidopsis mutants of sphingolipid fatty acid α-hydroxylases accumulate ceramides and salicylates. New Phytol. 196: 1086–1097. [DOI] [PubMed] [Google Scholar]
- Kosami K., Ohki I., Nagano M., Furuita K., Sugiki T., Kawano Y., Kawasaki T., Fujiwara T., Nakagawa A., Shimamoto K., Kojima C. (2014). The crystal structure of the plant small GTPase OsRac1 reveals its mode of binding to NADPH oxidase. J. Biol. Chem. 289: 28569–28578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi M., et al. (2007). Insights into the role of specific lipids in the formation and delivery of lipid microdomains to the plasma membrane of plant cells. Plant Physiol. 143: 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M., Bracha-Drori K., Sternberg H., Yalovsky S. (2002). A cell-specific, prenylation-independent mechanism regulates targeting of type II RACs. Plant Cell 14: 2431–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre B., Furt F., Hartmann M.A., Michaelson L.V., Carde J.P., Sargueil-Boiron F., Rossignol M., Napier J.A., Cullimore J., Bessoule J.J., Mongrand S. (2007). Characterization of lipid rafts from Medicago truncatula root plasma membranes: a proteomic study reveals the presence of a raft-associated redox system. Plant Physiol. 144: 402–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental I., Lingwood D., Grzybek M., Coskun U., Simons K. (2010). Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc. Natl. Acad. Sci. USA 107: 22050–22054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Liu P., Wan Y., Chen T., Wang Q., Mettbach U., Baluska F., Samaj J., Fang X., Lucas W.J., Lin J. (2012). A membrane microdomain-associated protein, Arabidopsis Flot1, is involved in a clathrin-independent endocytic pathway and is required for seedling development. Plant Cell 24: 2105–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberherr D., Thao N.P., Nakashima A., Umemura K., Kawasaki T., Shimamoto K. (2005). A sphingolipid elicitor-inducible mitogen-activated protein kinase is regulated by the small GTPase OsRac1 and heterotrimeric G-protein in rice 1[w]. Plant Physiol. 138: 1644–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D., Simons K. (2007). Detergent resistance as a tool in membrane research. Nat. Protoc. 2: 2159–2165. [DOI] [PubMed] [Google Scholar]
- Lingwood D., Simons K. (2010). Lipid rafts as a membrane-organizing principle. Science 327: 46–50. [DOI] [PubMed] [Google Scholar]
- Liu P., Li R.L., Zhang L., Wang Q.L., Niehaus K., Baluska F., Samaj J., Lin J.X. (2009). Lipid microdomain polarization is required for NADPH oxidase-dependent ROS signaling in Picea meyeri pollen tube tip growth. Plant J. 60: 303–313. [DOI] [PubMed] [Google Scholar]
- Löfgren H., Pascher I. (1977). Molecular arrangements of sphingolipids. The monolayer behaviour of ceramides. Chem. Phys. Lipids 20: 273–284. [DOI] [PubMed] [Google Scholar]
- Luttgeharm K.D., Kimberlin A.N., Cahoon E.B. (2016). Plant sphingolipid metabolism and function. Subcell. Biochem. 86: 249–286. [DOI] [PubMed] [Google Scholar]
- Marino D., Dunand C., Puppo A., Pauly N. (2012). A burst of plant NADPH oxidases. Trends Plant Sci. 17: 9–15. [DOI] [PubMed] [Google Scholar]
- Markham J.E., Li J., Cahoon E.B., Jaworski J.G. (2006). Separation and identification of major plant sphingolipid classes from leaves. J. Biol. Chem. 281: 22684–22694. [DOI] [PubMed] [Google Scholar]
- Markham J.E., Jaworski J.G. (2007). Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 21: 1304–1314. [DOI] [PubMed] [Google Scholar]
- Markham J.E., Lynch D.V., Napier J.A., Dunn T.M., Cahoon E.B. (2013). Plant sphingolipids: function follows form. Curr. Opin. Plant Biol. 16: 350–357. [DOI] [PubMed] [Google Scholar]
- Martinier A., et al. (2012). Cell wall constrains lateral diffusion of plant plasma-membrane proteins. Proc. Natl. Acad. Sci. USA 109: 12805–12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D., Shimamoto K. (2004). Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45: 490–495. [DOI] [PubMed] [Google Scholar]
- Moissoglu K., Kiessling V., Wan C., Hoffman B.D., Norambuena A., Tamm L.K., Schwartz M.A. (2014). Regulation of Rac1 translocation and activation by membrane domains and their boundaries. J. Cell Sci. 127: 2565–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongrand S., Morel J., Laroche J., Claverol S., Carde J.P., Hartmann M.A., Bonneu M., Simon-Plas F., Lessire R., Bessoule J.J. (2004). Lipid rafts in higher plant cells: purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem. 279: 36277–36286. [DOI] [PubMed] [Google Scholar]
- Mongrand S., Stanislas T., Bayer E.M., Lherminier J., Simon-Plas F. (2010). Membrane rafts in plant cells. Trends Plant Sci. 15: 656–663. [DOI] [PubMed] [Google Scholar]
- Morel J., Claverol S., Mongrand S., Furt F., Fromentin J., Bessoule J.J., Blein J.P., Simon-Plas F. (2006). Proteomics of plant detergent-resistant membranes. Mol. Cell. Proteomics 5: 1396–1411. [DOI] [PubMed] [Google Scholar]
- Nakashima A., Chen L., Thao N.P., Fujiwara M., Wong H.L., Kuwano M., Umemura K., Shirasu K., Kawasaki T., Shimamoto K. (2008). RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell 20: 2265–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M., Ihara-Ohori Y., Imai H., Inada N., Fujimoto M., Tsutsumi N., Uchimiya H., Kawai-Yamada M. (2009). Functional association of cell death suppressor, Arabidopsis Bax inhibitor-1, with fatty acid 2-hydroxylation through cytochrome b₅. Plant J. 58: 122–134. [DOI] [PubMed] [Google Scholar]
- Nagano M., Takahara K., Fujimoto M., Tsutsumi N., Uchimiya H., Kawai-Yamada M. (2012a). Arabidopsis sphingolipid fatty acid 2-hydroxylases (AtFAH1 and AtFAH2) are functionally differentiated in fatty acid 2-hydroxylation and stress responses. Plant Physiol. 159: 1138–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M., Uchimiya H., Kawai-Yamada M. (2012b). Plant sphingolipid fatty acid 2-hydroxylases have unique characters unlike their animal and fungus counterparts. Plant Signal. Behav. 7: 1388–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M., Ishikawa T., Ogawa Y., Iwabuchi M., Nakasone A., Shimamoto K., Uchimiya H., Kawai-Yamada M. (2014). Arabidopsis Bax inhibitor-1 promotes sphingolipid synthesis during cold stress by interacting with ceramide-modifying enzymes. Planta 240: 77–89. [DOI] [PubMed] [Google Scholar]
- Navarro-Lérida I., Sánchez-Perales S., Calvo M., Rentero C., Zheng Y., Enrich C., Del Pozo M.A. (2012). A palmitoylation switch mechanism regulates Rac1 function and membrane organization. EMBO J. 31: 534–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T., Hashimoto H., Kuwabara N., Akashi S., Hayashi K., Kojima C., Wong H.L., Kawasaki T., Shimamoto K., Sato M., Shimizu T. (2010). Structure of the N-terminal regulatory domain of a plant NADPH oxidase and its functional implications. J. Biol. Chem. 285: 1435–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D.M., Rentero C., Magenau A., Abu-Siniyeh A., Gaus K. (2011). Quantitative imaging of membrane lipid order in cells and organisms. Nat. Protoc. 7: 24–35. [DOI] [PubMed] [Google Scholar]
- Pascher I., Sundell S. (1977). Molecular arrangements in sphingolipids. The crystal structure of cerebroside. Chem. Phys. Lipids 20: 175–191. [DOI] [PubMed] [Google Scholar]
- Raffaele S., et al. (2009). Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21: 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche Y., Gerbeau-Pissot P., Buhot B., Thomas D., Bonneau L., Gresti J., Mongrand S., Perrier-Cornet J.M., Simon-Plas F. (2008). Depletion of phytosterols from the plant plasma membrane provides evidence for disruption of lipid rafts. FASEB J. 22: 3980–3991. [DOI] [PubMed] [Google Scholar]
- Shimizu T., Nakano T., Takamizawa D., Desaki Y., Ishii-Minami N., Nishizawa Y., Minami E., Okada K., Yamane H., Kaku H., Shibuya N. (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64: 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Plas F., Perraki A., Bayer E., Gerbeau-Pissot P., Mongrand S. (2011). An update on plant membrane rafts. Curr. Opin. Plant Biol. 14: 642–649. [DOI] [PubMed] [Google Scholar]
- Sorek N., Poraty L., Sternberg H., Bar E., Lewinsohn E., Yalovsky S. (2007). Activation status-coupled transient S acylation determines membrane partitioning of a plant Rho-related GTPase. Mol. Cell. Biol. 27: 2144–2154. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sorek N., Segev O., Gutman O., Bar E., Richter S., Poraty L., Hirsch J.A., Henis Y.I., Lewinsohn E., Jürgens G., Yalovsky S. (2010). An S-acylation switch of conserved G domain cysteines is required for polarity signaling by ROP GTPases. Curr. Biol. 20: 914–920. [DOI] [PubMed] [Google Scholar]
- Stanislas T., Bouyssie D., Rossignol M., Vesa S., Fromentin J., Morel J., Pichereaux C., Monsarrat B., Simon-Plas F. (2009). Quantitative proteomics reveals a dynamic association of proteins to detergent-resistant membranes upon elicitor signaling in tobacco. Mol. Cell. Proteomics 8: 2186–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski W.G., Zauber H., Erban A., Gorka M., Wu X.N., Schulze W.X. (2015). Cytoskeletal components define protein location to membrane microdomains. Mol. Cell. Proteomics 14: 2493–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi D., Kawamura Y., Uemura M. (2013). Detergent-resistant plasma membrane proteome to elucidate microdomain functions in plant cells. Front. Plant Sci. 4: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ternes P., Feussner K., Werner S., Lerche J., Iven T., Heilmann I., Riezman H., Feussner I. (2011). Disruption of the ceramide synthase LOH1 causes spontaneous cell death in Arabidopsis thaliana. New Phytol. 192: 841–854. [DOI] [PubMed] [Google Scholar]
- Thao N.P., Chen L., Nakashima A., Hara S., Umemura K., Takahashi A., Shirasu K., Kawasaki T., Shimamoto K. (2007). RAR1 and HSP90 form a complex with Rac/Rop GTPase and function in innate-immune responses in rice. Plant Cell 19: 4035–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townley H.E., McDonald K., Jenkins G.I., Knight M.R., Leaver C.J. (2005). Ceramides induce programmed cell death in Arabidopsis cells in a calcium-dependent manner. Biol. Chem. 386: 161–166. [DOI] [PubMed] [Google Scholar]
- Uemura M., Joseph R.A., Steponkus P.L. (1995). Cold acclimation of Arabidopsis thaliana (effect on plasma membrane lipid composition and freeze-induced lesions). Plant Physiol. 109: 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Imai H. (2011). Characterization of glucosylceramides in leaves of the grass family (Poaceae): Pooideae has unsaturated hydroxy fatty acids. Biosci. Biotechnol. Biochem. 75: 1838–1841. [DOI] [PubMed] [Google Scholar]
- Wewer V., Dombrink I., vom Dorp K., Dörmann P. (2011). Quantification of sterol lipids in plants by quadrupole time-of-flight mass spectrometry. J. Lipid Res. 52: 1039–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H.L., Pinontoan R., Hayashi K., Tabata R., Yaeno T., Hasegawa K., Kojima C., Yoshioka H., Iba K., Kawasaki T., Shimamoto K. (2007). Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 19: 4022–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.