Figure 5.
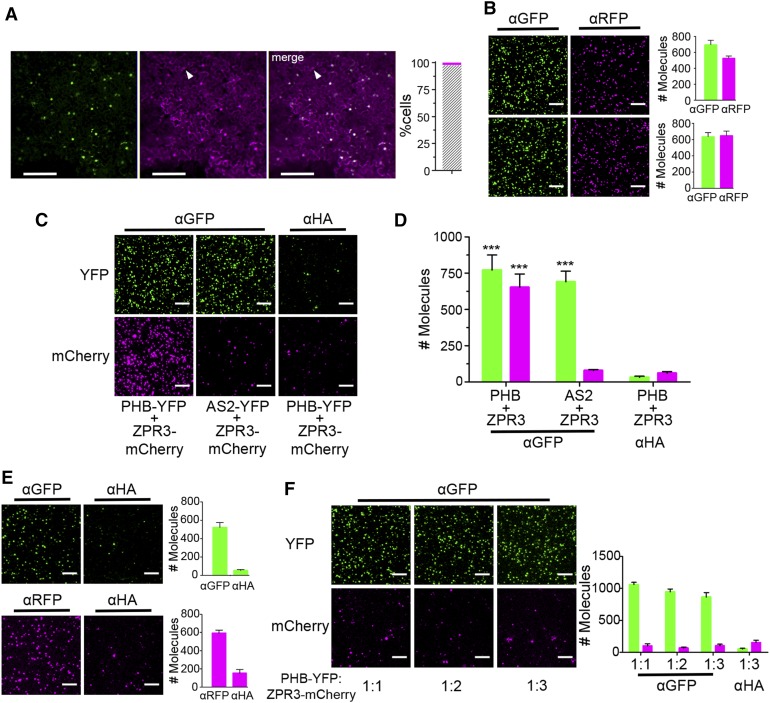
Detection of PHB-YFP and ZPR3-mCherry Coimmunoprecipitation via SiMPull.
(A) Fluorescence images of leaves infiltrated with a single Agrobacterium strain carrying 2xpro35S:PHB-YFP and pro35S:ZPR3-mCherry show roughly equivalent levels and near-perfect colocalization (gray hatched bar) of PHB-YFP and ZPR3-mCherry fluorescence 25 hpi. Arrowhead indicates unifluorescent cell.
(B) Representative TIRF images and IP spot counts from SiMPull using lysates coexpressing PHB-YFP and ZPR3-mCherry (top) or AS2-YFP and ZPR3-mCherry (bottom) show proteins are at single-spot resolution and accumulate to near-equivalent levels.
(C) and (D) SiMPull using PHB-YFP + ZPR3-mCherry lysate shows PHB-YFP IP and ZPR3-mCherry co-IP spot numbers are comparably enriched over background, whereas in SiMPull using AS2-YFP + ZPR3-mCherry negative control lysate, ZPR3-mCherry co-IP spots are indistinguishable from background.
(E) SiMPull of PHB-YFP (top) and ZPR3-mCherry (bottom) on lysates from separate Agrobacterium infiltrations with pro35S:PHB-YFP and pro35S:ZPR3-mCherry, respectively, identifies dilutions that yield equivalent IP spot numbers.
(F) SiMPull using these calibrated PHB-YFP and ZPR3-mCherry lysates mixed in vitro at different ratios demonstrates PHB-YFP does not coimmunoprecipitate ZPR3-mCherry under these conditions. Green, PHB-YFP or AS2-YFP; magenta, ZPR3-mCherry. Spot densities (means ± se) are calculated from at least three independent biological replicates. Student’s t test: ***P < 0.001.
Bars = 150 µm in (A) and 5 µm in (B) and (C).