Genomic imprinting is a rapidly evolving phenomenon in plants; however, for those genes where imprinting conveys an advantage, imprinted expression is maintained over long evolutionary timescales.
Abstract
Genomic imprinting is an epigenetic phenomenon occurring in mammals and flowering plants that causes genes to adopt a parent-of-origin-specific mode of expression. While the imprinting status of genes is well conserved in mammals, clear estimates for the degree of conservation were lacking in plants. We therefore analyzed the genome-wide imprinting status of Capsella rubella, which shared a common recent ancestor with Arabidopsis thaliana ∼10 to 14 million years ago. However, only ∼14% of maternally expressed genes (MEGs) and ∼29% of paternally expressed genes (PEGs) in C. rubella were commonly imprinted in both species, revealing that genomic imprinting is a rapidly evolving phenomenon in plants. Nevertheless, conserved PEGs exhibited signs of selection, suggesting that a subset of imprinted genes play an important functional role and are therefore maintained in plants. Like in Arabidopsis, PEGs in C. rubella are frequently associated with the presence of transposable elements that preferentially belong to helitron and MuDR families. Our data further reveal that MEGs and PEGs differ in their targeting by 24-nucleotide small RNAs and asymmetric DNA methylation, suggesting different mechanisms establishing DNA methylation at MEGs and PEGs.
INTRODUCTION
Genomic imprinting is an epigenetic phenomenon rendering gene expression in a parent-of-origin-dependent manner, thus violating the predictions of Mendel’s rules. Imprinting has evolved independently in placental mammals and flowering plants (Pires and Grossniklaus, 2014), and while it occurs in all tissue types in the former (Prickett and Oakey, 2012), it predominantly occurs in the endosperm in the latter (Hsieh et al., 2011; Luo et al., 2011; Gehring et al., 2011). The endosperm is an ephemeral triploid nutritive tissue supporting embryo growth, similar to the developmental role of the placenta in mammals. Endosperm development is initiated after fertilization of the diploid maternal central cell by a haploid sperm cell and it is consumed during embryo development or after germination (Li and Berger, 2012). Genomic imprinting has likely evolved as a consequence of transposable element (TE) insertions and the resulting host response to silence those elements (Suzuki et al., 2007; Pask et al., 2009; Jiang and Köhler, 2012). In agreement with this notion, differential DNA methylation is an important regulator of imprinted gene expression (Hsieh et al., 2009, 2011; Gehring et al., 2011; Pignatta et al., 2014). Genes that are controlled by imprinting mechanisms are targets for natural selection, with unequal effects for maternally and paternally derived alleles. Intragenomic conflict over the distribution of resources from the mother to the offspring provides one possible explanation for different selective forces acting on imprinted genes (Moore and Haig, 1991). Natural selection for increased offspring fitness by increasing the probability of expressing the fitter of the two parental alleles could similarly explain why imprinted expression is maintained (Wolf and Hager, 2006). In mammals, the imprinting status of most imprinted genes is conserved between mouse and human and some are imprinted even in marsupials (Murphy and Jirtle, 2003; Suzuki et al., 2005), indicating the significance of genomic imprinting for mammalian evolution. To which extent the imprinting status of genes has been conserved in plants is difficult to estimate based on available studies. The genome-wide imprinting status in plants has been identified in a range of different species, namely, Arabidopsis thaliana (Hsieh et al., 2011; Wolff et al., 2011; Gehring et al., 2011; Pignatta et al., 2014), rice (Oryza sativa; Luo et al., 2011), maize (Zea mays; Waters et al., 2013), castor bean (Ricinus communis; Xu et al., 2014), and sorghum (Sorghum bicolor; Zhang et al., 2016). The overlap of imprinted genes identified in these different studies is rather low, ranging from 13% conserved imprinted genes between maize and rice (Zhang et al., 2016) to 4% conserved imprinted genes between Arabidopsis and castor bean (Xu et al., 2014). However, the long divergence time of rice and maize (∼50 million years ago [Mya]; Lai et al., 2004) as well as of Arabidopsis and castor bean (∼110 Mya; Zheng et al., 2013) make direct inference about the conservation of the imprinting status in plants difficult. While maize and sorghum recently diverged (around ∼12 Mya; Swigonová et al., 2004), they share only 29% conserved imprinted genes. Nevertheless, despite their recent divergence, maize has undergone an allopolyploidization event and an approximate 3-fold genome expansion since its divergence from sorghum (Gaut and Doebley, 1997; Paterson et al., 2009), complicating direct inference about the degree of conservation of imprinted genes in both species. In this study, we set out to test the imprinting status in two closely related dicot species, Arabidopsis and Capsella rubella, which belong to the same family, share a common recent ancestor ∼10 to 14 Mya (Mitchell-Olds, 2001; Koch and Kiefer, 2005), and have similar transposable element frequency and density (Slotte et al., 2013), allowing us to specifically test the conservation of imprinting over short evolutionary time periods.
RESULTS
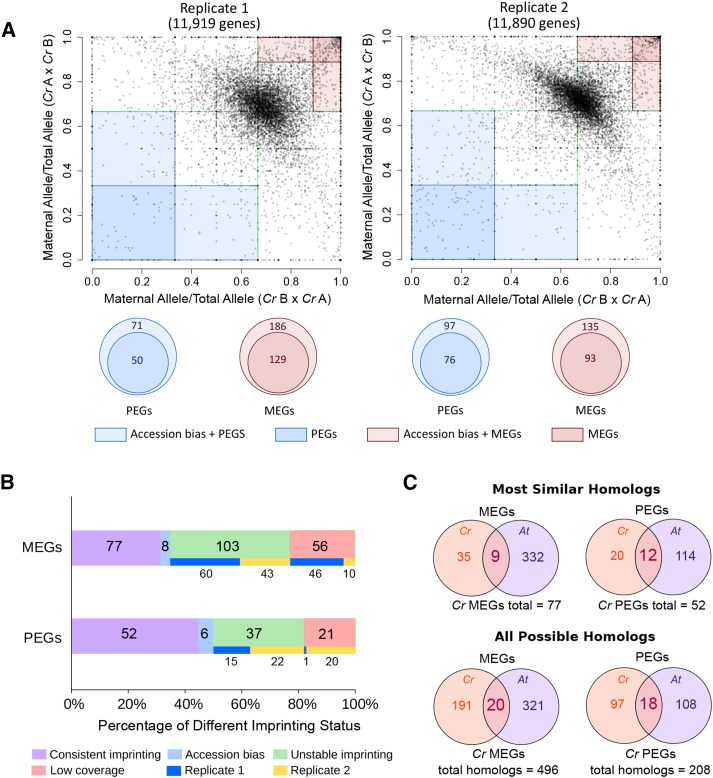
To analyze the imprinting status in C. rubella, we performed reciprocal crosses between two accessions that differed in 168,634 small nucleotide polymorphisms (SNPs) in 15,293 protein coding genes, which corresponds to ∼58% of the genes present in the C. rubella genome (Slotte et al., 2013). Analysis of hybrid seed development did not reveal any postzygotic barrier or developmental defect in comparison to self-fertilized seeds (Supplemental Figures 1A to 1C). We deep sequenced endosperm isolated from reciprocally crossed C. rubella accessions at 6 d after pollination (DAP) in two biological replicates. At 6 DAP, the majority of seeds were at the late heart to early torpedo stage of development (Supplemental Figure 1A), which is comparable to the developmental stage previously analyzed in imprinting studies of Arabidopsis (Hsieh et al., 2011; Gehring et al., 2011; Pignatta et al., 2014). A large number of reads (118 million and 137 million) were recovered for each of the cross combinations and analyzed for allelic expression patterns (for details, see Methods and Supplemental Table 1). The number of reads that mapped to each allele was summed across all SNPs and only annotated genes that had 20 informative parental reads were analyzed (12,641 genes). Maternally expressed imprinted genes (MEGs) preferentially express the maternal allele in both directions of a cross, whereas paternally expressed imprinted genes (PEGs) preferentially express the paternal allele in both directions of a cross. Genes that showed parental-specific expression only in one direction of the cross but were biallelically expressed in the reciprocal cross were categorized as accession-biased genes. Following previously applied strategies (Waters et al., 2013; Pignatta et al., 2014), MEGs and PEGs were identified by implementing a combination of statistical significance and proportion filters. Specifically, we defined MEGs and PEGs as having significant allelic bias (χ2 P value < 0.01) from the expected 2:1 ratio of maternal to paternal transcripts and ≥89% of transcripts from the maternal allele (MEGs) or ≥67% of the transcripts from the paternal allele (PEGs) (red and blue shaded areas in Figure 1A, respectively) in both replicates and both directions of a reciprocal cross. Based on these criteria, we identified 50 MEGs and 29 PEGs (Supplemental Data Set 1). Several potentially imprinted genes passed all stringency criteria in one replicate, but while having a clear allelic bias, they did not pass all criteria in the second replicate. We therefore also classified genes as being imprinted if they met all criteria for an imprinted gene in one of the biological replicates, and in the second replicate had an informative read count of at least 10 for the respective cross and had either a significant allelic bias (χ2 P value < 0.01 or ≥80% of transcripts from the maternal allele [MEGs] or ≥50% of the transcripts from the paternal allele [PEGs]). Additional filtering criteria were applied to MEGs to remove genes that might exhibit maternal bias due to contamination of maternally derived seed coat tissues. We generated RNA-seq data from isolated seeds of the parental accessions and compared the transcriptome profiles of seed and endosperm tissues (see Methods for details). Genes with decreased expression level in the endosperm libraries compared with the whole seed libraries were considered as maternal tissue contaminants. To validate this method, we identified the 50 most similar C. rubella homologs to genes that were highly expressed in embryo and seed coat in Arabidopsis (Belmonte et al., 2013). Of those, we identified between two and seven genes in our libraries, allowing us to estimate that the maximal false discovery rate for MEGs is 14% (Supplemental Data Set 2). Applying these criteria, the total number of imprinted genes in both replicates was 77 MEGs and 52 PEGs (Supplemental Data Set 2; Figure 1B). We also identified 14 accession-biased genes that showed parental-specific expression only in one direction of the cross, but were biallelically expressed in the reciprocal cross (Supplemental Data Set 3). We validated the imprinting status of 10 randomly selected MEGs and PEGs and confirmed the imprinting status of 8 MEGs and 10 PEGs, while two MEGs were imprinted only in one direction of the cross (Supplemental Figure 2 and Supplemental Data Set 1). We further tested our applied thresholds by performing a principal component analysis of imprinted genes and those genes that had been identified as imprinted in Arabidopsis but had not been identified as imprinted in C. rubella based on our criteria (Supplemental Data Set 5 and Supplemental Figure 3A). This analysis revealed that C. rubella MEGs and PEGs formed largely distinct clusters that were clearly separated from nonimprinted control genes. The majority of genes that were imprinted in Arabidopsis but that had been classified as nonimprinted in C. rubella clustered together with nonimprinted control genes. A subset of those genes showed a parental bias and very few of them clustered together with imprinted genes. Those genes have not been classified as imprinted due to low read counts. Gene expression levels between C. rubella MEGs, PEGs, and genes that are imprinted in Arabidopsis but that have been classified as nonimprinted in C. rubella were not significantly different (Mann-Whitney pairwise test, P value > 0.05) (Supplemental Data Set 4 and Supplemental Figure 3B), making it unlikely that different expression levels of imprinted genes in both species biased their identification. Together, these data reveal that our selection criteria reliably identified imprinted genes in C. rubella.
Figure 1.
Only a Small Subset of Imprinted Genes Is Conserved between C. rubella and Arabidopsis.
(A) Allele‐specific expression analysis. The proportion of maternal transcripts in both reciprocal hybrids was plotted for all genes with available reads for both replicates.
(B) Imprinting stability in both replicates. Consistent imprinting: consistently imprinted genes in both biological replicates. Accession biased: parental‐specific expression only in one direction of the cross but not in the other in both replicates. Unstable imprinting: genes that were imprinted only in one of the two replicates. Low coverage: genes with insufficient read counts in one of the two replicates.
(C) Overlap of most similar (upper charts) and all possible (lower charts) Arabidopsis homologs of C. rubella imprinted genes with Arabidopsis imprinted genes (Hsieh et al., 2011; Gehring et al., 2011; Pignatta et al., 2014). Cr, C. rubella; At, Arabidopsis.
Comparison between C. rubella and Arabidopsis Imprinted Genes Reveals Low Imprinting Conservation
We compared the identified C. rubella imprinted genes with those imprinted genes previously identified in Arabidopsis (Hsieh et al., 2011; Wolff et al., 2011; Gehring et al., 2011; Pignatta et al., 2014). We considered a gene to be imprinted in Arabidopsis if it was identified in at least one of the previously published studies using Columbia and Landsberg erecta accessions (Hsieh et al., 2011; Wolff et al., 2011; Gehring et al., 2011; Pignatta et al., 2014). For almost all imprinted C. rubella genes, we identified close homologs in Arabidopsis (76 out of 77 MEGs and 52 out of 52 PEGs; Supplemental Data Set 5). Of those, 9 MEGs and 12 PEGs were imprinted in both species (Supplemental Data Set 5), while 35 MEGs and 20 PEGs of C. rubella homologs in Arabidopsis were biallelically expressed (1.5 ≤ M/P ≤ 2.5). For homologs of 18 MEGs and 9 PEGs, sufficient read counts were not detectable in Arabidopsis (Supplemental Data Set 5), suggesting that they are not substantially expressed during endosperm development and likely have different functional roles compared with their homologs in C. rubella. The imprinting status of 13 C. rubella MEGs and 10 PEGs homologs could not be tested in Arabidopsis because they lack SNPs. Thus, out of 63 C. rubella MEGs and 42 PEGs that had close homologs and SNPs in Arabidopsis, only 9 MEGs (14.3%) and 12 PEGs (28.6%) were also imprinted in Arabidopsis, revealing a low degree of imprinting conservation. We were not able to test the imprinting status of the Arabidopsis homologs of MEDEA (MEA; Carubv10012183m) and FERTILIZATION INDEPENDENT SEED2 (FIS2; Carubv10024660m) in C. rubella due to low coverage for MEA and no SNPs available for FIS2. Interestingly, the homologs of the Arabidopsis MEG FLOWERING WAGENINGEN (FWA; Carubv10007084m) and the PEG PHERES1 (PHE1; Carubv10001714m) switched their parental preference in C. rubella, with FWA being a PEG and PHE1 being a MEG.
We extended the homology search of imprinted C. rubella genes in Arabidopsis by including all homologous genes with an e-value ≤ 1e-5. This comparison revealed that C. rubella had 20 MEGs (9.5%) and 18 PEGs (15.6%) with their homologs being imprinted in Arabidopsis (Figure 1C, lower charts). The substantially increased number of conserved imprinted genes between both species when including distant homologs suggests that the imprinting status rapidly changes over evolutionary time; however, for those genes where imprinting may convey an advantage, it will be maintained.
Conserved and Nonconserved Imprinted Genes Have Different Functional Roles
One prediction of the hypothesis that imprinting of some genes confers an advantage is that conserved and nonconserved imprinted genes belong to different functional categories. We tested this prediction by analyzing the Gene Ontology (GO) enrichment for conserved imprinted genes that were imprinted in C. rubella and had imprinted homologs in Arabidopsis (Figure 1C, lower panel; Supplemental Table 2) and nonconserved imprinted genes that were only found imprinted in one species but did not share imprinted homologs with the other. There was indeed a clear difference in GO enrichments between conserved and nonconserved MEGs and PEGs; while conserved MEGs were significantly enriched for genes connected to the regulation of transcription and metabolism (Supplemental Table 2), nonconserved MEGs in C. rubella and Arabidopsis did not show significant enrichment for specific functional categories. Conserved PEGs were enriched for genes related to chromatin organization, while nonconserved PEGs in Arabidopsis were enriched for genes related to transcriptional regulation (Supplemental Table 2). Nonconserved PEGs in C. rubella were not significantly enriched for specific functional categories. Together, this analysis reveals that conserved and nonconserved MEGs and PEGs have different functional roles, which may be a consequence of different selection pressures acting on both classes of imprinted genes.
We addressed the question whether the imprinting status of those genes that are commonly imprinted in Arabidopsis and C. rubella is conserved in other species that have been investigated thus far (Luo et al., 2011; Waters et al., 2013; Xu et al., 2014). We identified several genes commonly imprinted in Arabidopsis, C. rubella, and either rice (Oryza sativa) or maize (Supplemental Figure 4). Among those, the conserved imprinted genes VARIANT IN METHYLATION1 (VIM1), VIM3, VIM5, VIM6, and PICKLE RELATED2 have functional roles in chromatin modification (Woo et al., 2008; Aichinger et al., 2009), in agreement with gross changes in chromatin structure occurring after fertilization (She and Baroux, 2014). Imprinting of the auxin biosynthesis gene TAA1 is also conserved between Arabidopsis, C. rubella, and monocots, suggesting that the mechanism coupling endosperm development and fertilization via the paternal expression of TAA1 is evolutionarily conserved (Figueiredo et al., 2015).
Conserved PEGs Are Undergoing Adaptive Evolution
To test whether conserved and nonconserved imprinted genes are under different selection pressure, we compared neutral (4-fold synonymous) to putatively selected polymorphisms (0-fold nonsynonymous) to estimate the parameters of a gamma-distribution of negative fitness effect (DFE) and the proportion of adaptive substitutions at selected sites. To obtain sufficient numbers of polymorphisms for this analysis, we used polymorphism data generated by genome resequencing of 12 samples from separate populations of Capsella grandiflora covering the geographical range of the species. As the vast majority of polymorphisms in C. rubella is shared with C. grandiflora (Foxe et al., 2009), we expect results to be representative for C. rubella. Control genes to imprinted genes were selected at random from nonimprinted genes that were expressed in the endosperm and harbored sufficient SNPs for imprinting analyses.
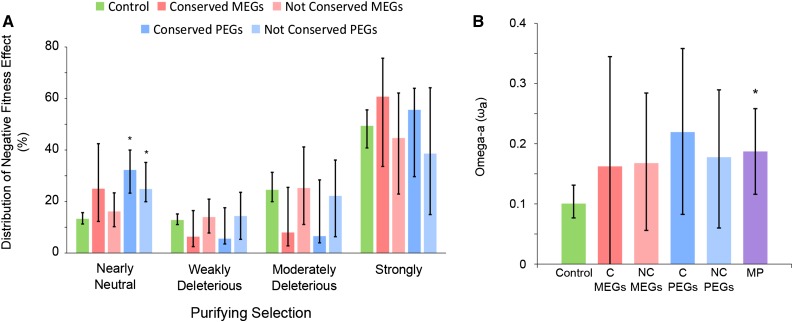
We found that PEGs as well as conserved PEGs had a significantly higher proportion of nearly neutral nonsynonymous variants compared with the control genes (Figure 2A), indicating that PEGs are experiencing weaker purifying selection. The differences between MEGs and conserved MEGs were not statistically significant (Figure 2A). When analyzing the rate of adaptive substitution relative to the rate of neutral evolution (ωa) as a robust measure of adaptive evolution (Eyre-Walker and Keightley, 2009; Gossmann et al., 2010), ωa was higher in MEGs and PEGs compared with the control and even higher in conserved PEGs compared with PEGs, while this trend did not hold for conserved MEGs (Figure 2B). Though due to low sample size the differences were not significant, the differences compared with the control were significant when MEGs and PEGs were analyzed together (Figure 2B). Together, these data suggest that a subset of MEGs and PEGs is under positive selection and particularly conserved PEGs may be undergoing elevated adaptive evolution.
Figure 2.
Imprinted Genes Show Signs of Positive Selection.
(A) DFE for different groups of C. rubella imprinted genes. Whiskers represent 95% confidence interval. Asterisks mark significant changes compared with the control (P value < 0.05). Significance is based on comparison of 200 bootstrapped data sets.
(B) Rate of adaptive substitution relative to the rate of neutral evolution (ωa). Whiskers represent 95% confidence interval. Asterisks mark significant changes compared with the control (P value < 0.05). Significance is based on comparison of 200 bootstrapped data sets. Conserved (C) MEGs and PEGs correspond to genes that are imprinted in C. rubella and have distant imprinted homologs in Arabidopsis; nonconserved (NC) MEGs and PEGs correspond to genes that are exclusively imprinted in C. rubella; control genes are nonimprinted genes in C. rubella with similar expression levels as imprinted genes and harbored sufficient SNPs for imprinting analyses. MP corresponds to MEGs and PEGs joined together regardless the conservation.
Transposable Elements Are Enriched in the Proximity of PEGs
TE insertions appear to be an important driver of imprinted gene expression (Suzuki et al., 2007; Pask et al., 2009; Pignatta et al., 2014) and in agreement with this notion, around one-third of imprinted genes in Arabidopsis are flanked by TEs (Jiang and Köhler, 2012; Pignatta et al., 2014). We addressed the question of whether the difference of genes imprinted in one species but not in the other correlates with the presence of TE insertions in the vicinity of those genes. Using RepeatMasker (http://www.repeatmasker.org/), we scanned the proximity and the gene body of imprinted genes for the presence of repeats as a measure for the presence of TEs (Lerat, 2010).
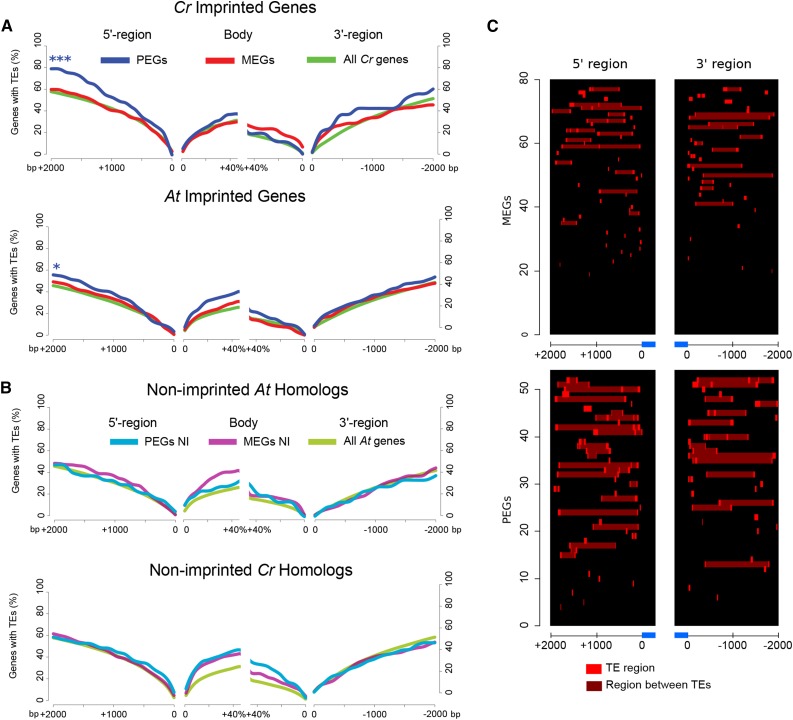
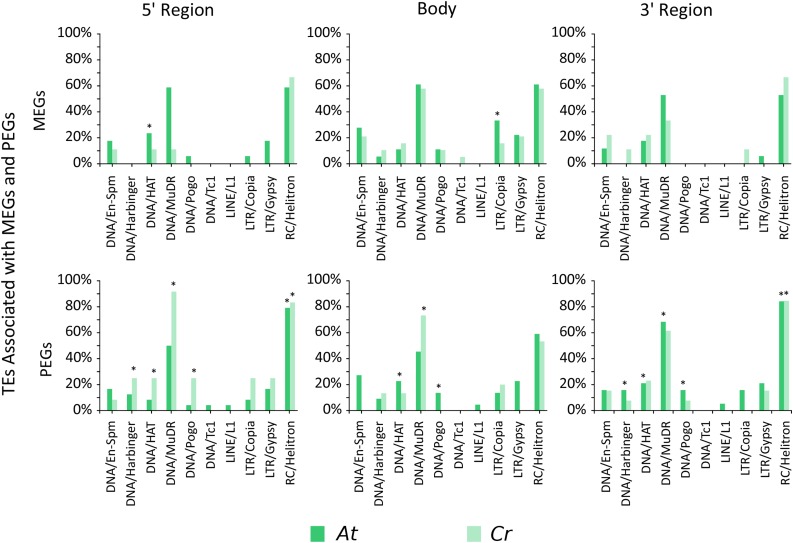
We observed a significant enrichment of TEs in upstream regions of C. rubella PEGs (Figure 3A, upper panel; Supplemental Data Set 6), mirroring the enrichment of TEs in Arabidopsis PEGs (Figure 3A, lower panel) (Pignatta et al., 2014). Fewer MEGs were associated with TEs compared with PEGs (Figures 3A and 3C), supporting previous observations in Arabidopsis (Pignatta et al., 2014). We analyzed the presence of TEs in those Arabidopsis genes that were nonimprinted homologs of imprinted C. rubella genes. The nonimprinted status of Arabidopsis PEG homologs significantly correlated with the absence of TEs in the flanking regions of those genes (Figure 3B, upper panel). Similarly, nonimprinted C. rubella homologs of imprinted Arabidopsis PEGs were not significantly associated with TEs (Figure 3B, lower panel). Together, these data reveal that the presence of TEs is closely associated with the imprinting status of PEGs in C. rubella and Arabidopsis and that the absence of TEs correlates with the nonimprinted status of the corresponding homologs. Thus, transposon dynamics can explain the low conservation level of imprinting among species, consistent with TEs being highly dynamic and driving rapid genome changes (Tenaillon et al., 2010). We found that helitrons and MuDR elements were most commonly found in the gene body and surrounding regions of PEGs, reflecting their abundance in the Arabidopsis and C. rubella genomes and their preferred association with differentially methylated regions (Pignatta et al., 2014) (Figure 4; Supplemental Data Set 7). Associated TEs found in and around PEGs are consistent with previous findings made in Arabidopsis (Wolff et al., 2011; Pignatta et al., 2014). In a separate analysis we also tested for the presence of imprinted TEs and identified 17 imprinted TEs that were all maternally expressed (Supplemental Data Set 8). However, those TEs were not flanking imprinted genes.
Figure 3.
TEs Preferentially Accumulate in the Proximity of Imprinted Genes.
(A) Top panel: percentage of C. rubella MEGs, PEGs, and all C. rubella (Cr) genes with TEs. Significance of TE enrichment was tested using a hypergeometric test. Lower panel: percentage of Arabidopsis MEGs, PEGs, and all Arabidopsis (At) genes with TEs. Significance of TE enrichment was tested using a hypergeometric test. ***P ≤ 0.001 and *P ≤ 0.05.
(B) Top panel: percentage of nonimprinted (NI) Arabidopsis homologs of C. rubella imprinted genes with TEs versus all Arabidopsis genes. Lower panel: percentage of nonimprinted (NI) C. rubella homologs of Arabidopsis imprinted genes with TEs versus all C. rubella genes.
(C) Distribution of TEs in the vicinity of C. rubella MEGs and PEGs. “Region between TEs” refers to a region that is located between two or more TEs.
Figure 4.
Presence of TEs Belonging to Defined TE Classes in the 5′ Coding and 3′ Region of Imprinted Genes in C. rubella and Arabidopsis.
The plot shows which potential TE family is associated with significantly enriched simple repeats in the flanking region (2 kb) and coding region of imprinted genes. Significantly enriched simple repeats in MEGs and PEGs were associated with certain TE families using Arabidopsis TE families as reference model. Significance of enrichment of certain TE families with imprinted genes was tested using a hypergeometric test.
Different Methylation Patterns at MEGs and PEGs
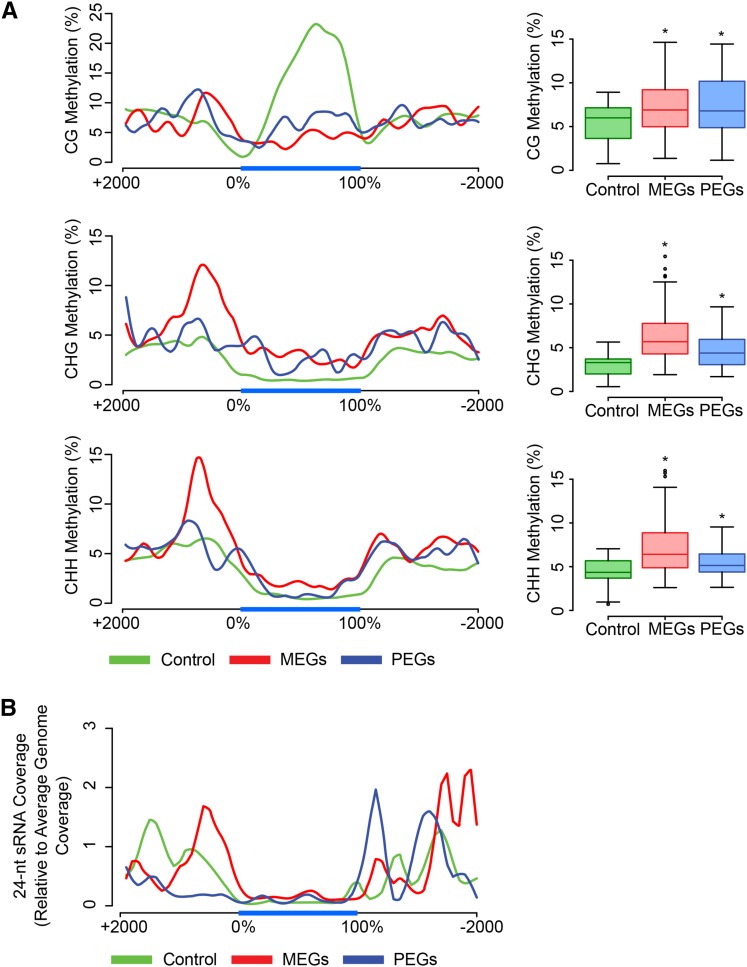
Differential DNA methylation of parental alleles is an important regulator of imprinted gene expression (Gehring et al., 2009; Hsieh et al., 2009; Ibarra et al., 2012). In agreement with this notion, we found MEGs and PEGs to be flanked by DNA methylated regions (Figure 5A; Supplemental Figure 5). Gene body CG methylation of all genes was considerable higher compared with those found in MEGs and PEGs, consistent with the fact that gene body CG methylation correlates with transcriptional activity (Zilberman et al., 2007) and many imprinted genes are not expressed in vegetative tissues (Wolff et al., 2011; Waters et al., 2013). Strikingly, the promoter regions of MEGs were marked by increased methylation in CHG and CHH context compared with PEGs and all genes (Figure 5A). Many euchromatic TEs and related sequences are targeted by RNA-directed DNA methylation, mediated by 24-nucleotide small RNAs (sRNAs) (Law and Jacobsen, 2010). Consistently, the promoter regions of MEGs accumulated high levels of 24-nucleotide sRNAs (Figure 5B), in agreement with previous studies (Calarco et al., 2012; Pignatta et al., 2014). The promoter regions of MEGs were not significantly enriched for TEs (Figure 3A), contrasting with the accumulation of 24-nucleotide sRNAs in those regions. However, most of the 24-nucleotide sRNAs targeting the promoter regions of MEGs were mapped at multiple locations in the genome (Figure 6), suggesting that they are generated from distantly located TEs in the genome and targeting the promoter regions of MEGs in trans. Promoter regions of PEGs were enriched for CG methylation but had substantially lower levels of CHG and CHH methylation, consistent with low level of 24-nucleotide sRNAs (Figure 5).
Figure 5.
DNA Methylation and 24‐Nucleotide sRNAs Are Enriched in Flanking Regions of C. rubella Imprinted Genes.
(A) Left panels: average CG, CHG, and CHH methylation in C. rubella in 5′ upstream (2 kb), transcribed, and 3′ downstream (2 kb) regions of PEGs (blue), MEGs (red), and control (green) genes. Right panel: differences between methylation level between imprinted genes and control genes in 0 to ±1-kb flanking regions. Asterisks mark significant difference (Wilcox‐Mann‐Whitney Test, P value < 0.05). Methylation data have been used from Seymour et al. (2014).
(B) The 24‐nucleotide sRNA densities in 5′ upstream (2 kb), transcribed, and 3′ downstream (2 kb) regions of PEGs, MEGs, and all genes.
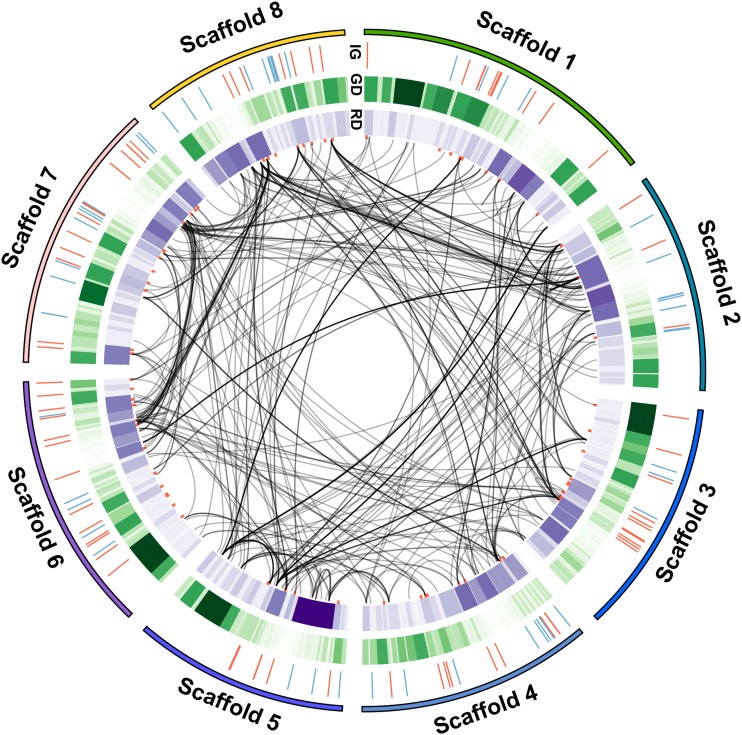
Figure 6.
The 24-Nucleotide sRNAs from the 5′ Upstream Regions of C. rubella MEGs Map to Multiple Positions in the Genome.
Darker color reflects increased density. Connecting black lines in the middle of the ideogram represent positions where the 24-nucleotide sRNAs can be mapped. Red dots in the most inner circle represent MEGs to aid visualization. Mapping of extracted 24-nucleotide sRNAs was done by BLAT using an optimized mapping score set to 10 to accommodate 24-nucleotide alignment. IG, imprinted genes; blue strokes, PEGs; red strokes, MEGs; GD, gene density; RD, repeat element density.
As a consequence of DNA methylation being enriched in the vicinity of MEGs and PEGs, we expected that imprinted genes should have a larger minimal distance to neighboring genes compared with the genome-wide average. We tested this hypothesis by scanning the occurrence of neighboring genes surrounding MEGs or PEGs (±500 bp from transcriptional start or stop). As predicted, the likelihood of finding neighboring genes for MEGs and PEGs was significantly lower (hypergeometric test, P value < 0.05) compared with all genes in the same species, with PEGs being the most isolated genes (Supplemental Figure 6).
DISCUSSION
Our data reveal that the accumulation of helitrons and MuDR elements in the vicinity of imprinted genes is a conserved feature in Arabidopsis and C. rubella. Strikingly, despite this conservation, less than one-third of orthologous genes are imprinted in both species, revealing that genomic imprinting is a highly dynamic process. We could show that the imprinting status correlates with the presence of TEs and, conversely, the absence of TEs with the nonimprinted status of genes, connecting the dynamic behavior of imprinting with the dynamics of TEs. Thus, it seems most likely that imprinted expression of genes is a consequence of epigenetic mechanisms targeting TEs. Interestingly, the same degree of imprinting conservation was reported for sorghum and maize (Zhang et al., 2016) that diverged at a similar time as Arabidopsis and C. rubella (Mitchell-Olds, 2001; Swigonová et al., 2004; Koch and Kiefer, 2005). Thus, despite extensive TE proliferation that occurred after divergence of sorghum and maize (Gaut and Doebley, 1997, Paterson et al., 2009), a similar number of conserved imprinted genes was maintained between both species, as in Arabidopsis and C. rubella, which also maintained a similar TE frequency and density after their divergence (Slotte et al., 2013). Importantly, genes commonly imprinted in Arabidopsis and C. rubella were enriched for specific functional categories that are also enriched when comparing conserved imprinted genes between the distantly related species Arabidopsis and maize (Waters et al., 2013), while nonconserved C. rubella imprinted genes did not share functional categories. This suggests that genomic imprinting of a small fraction of genes conveys an advantage and is therefore maintained over long evolutionary time scales, while in most instances it is either neutral or even disadvantageous and therefore rapidly purged. Alternatively, genetic conflict could be an important factor driving the rapid turnover of imprinted genes. Both Arabidopsis and C. rubella became self-compatible very recently during evolution (Bechsgaard et al., 2006; Foxe et al., 2009; Guo et al., 2009). Thus, both species were self-incompatible most of the time after the split from a common ancestor ∼10 to 14 Mya (Mitchell-Olds, 2001; Koch and Kiefer, 2005). Therefore, the large number of imprinted genes identified in Arabidopsis and C. rubella could be a consequence of a recent past as outcrossing species, while the shift of mating system could have accelerated the loss of conserved imprinted genes. Consistent with previous findings in monocots (Waters et al., 2013), conserved imprinted genes in Arabidopsis and C. rubella had increased ω values compared with nonconserved imprinted genes, suggesting that these genes are responsive to genomic conflict. While genomic conflict is not expected to play a substantial role in self-fertilizing species, the recent shift of mating type together with the substantial rate of outcrossing observed in Arabidopsis (Bomblies et al., 2010) provide an explanation why genes with potential functions in genomic conflict remain imprinted in self-fertilizing species. Nevertheless, recent work revealed that imprinted genes also play an important role in stress response (Sanchez and Paszkowski, 2014); thus, the functional role of some imprinted genes may be concealed only under certain stress conditions. Supporting this notion, dynamic DNA methylation changes are part of the plant-induced immune response (Dowen et al., 2012; Yu et al., 2013) and may therefore also affect the regulation of imprinted genes that are generally associated with differentially methylated regions (Gehring et al., 2009; Hsieh et al., 2009; Ibarra et al., 2012; Pignatta et al., 2014). Therefore, conserved imprinted genes may play a role outside the endosperm during plant stress response, an attractive hypothesis that remains to be thoroughly tested.
Our data further reveal that MEGs and PEGs differ in their targeting by 24-nucleotide sRNAs; while we found 24-nucleotide sRNAs targeting the promoter regions of MEGs, there was no substantial enrichment of 24-nucleotide sRNAs at PEGs. Consistently, CHH methylation, which is dependent on 24-nucleotide sRNAs (Law and Jacobsen, 2010), was mainly detected in the promoter regions of MEGs, but not PEGs. By contrast, the promoter regions of both MEGs and PEGs were marked by symmetric CG methylation. Maintenance of CG methylation can occur independently of 24-nucleotide sRNAs (Bond and Baulcombe, 2014), suggesting that different mechanisms function in maintaining DNA methylation on MEG- and PEG-associated TEs. Interestingly, in Arabidopsis the majority of evolutionarily old repeats corresponds to putative remnants of helitron TEs (Maumus and Quesneville, 2014) and helitrons associate with the majority of PEGs (Figure 4). In contrast to young repeats, which almost all overlap with 24-nucleotide sRNAs, only about half of old repeats are associated with 24-nucleotide sRNAs (Maumus and Quesneville, 2014), providing a possible explanation for the difference in 24-nucleotide sRNAs associating with flanking regions of MEGs and PEGs.
METHODS
Plant Material
The two Capsella rubella accessions used in this study were collected in Tuscany, Italy (Cr48.2-1 or Cr A) and Trapeza, Greece (Cr75.2-3 or Cr B) (St. Onge et al., 2011; Slotte et al., 2013). After stratification for 2 d in the dark at 4°C, seedlings were grown in a growth room under long-day photoperiod (16 h light and 8 h darkness) at 22°C light and 20°C darkness temperature and a light intensity of 110 μE from a Osram FQ 24W/840 HQ Constant Lumilux Cool White light source. Seedlings were transferred to pots and plants were grown in a growth chamber at 60% humidity and daily cycles of 16 h light at 21°C and 8 h darkness at 18°C. Designated female partners were emasculated, and the pistils were hand-pollinated 2 d after emasculation. Endosperm was isolated from 6 DAP seeds by manual dissection. For each library 300 seeds were dissected.
Microscopy
Clearing analysis was performed as previously described in Roszak and Köhler (2011).
DNA and RNA Isolation and Library Preparation
DNA was extracted from 200 mg of pooled leaves of 12 individual plants using the DNeasy Plant Mini Kit (Qiagen) and 2 μg of sheared DNA was used for library preparation following protocols of the TruSeq DNA sample preparation kit (Illumina).
RNA from endosperm was extracted using the RNAqueous kit with Plant RNA Isolation Aid (Ambion) and preserved in RNAlater (Sigma-Aldrich). RNA-seq libraries were prepared using the TruSeq RNA kit (Illumina). Libraries were sequenced at the SciLife Laboratory on an Illumina HiSeq2000 in 100-bp paired-end fashion.
sRNA Data and DNA Methylation Analysis
sRNA data of C. rubella accession (Cr1GR1) inflorescence tissues was generated, filtered, and mapped as previously published (Steige et al., 2015), using the C. rubella genome (Steige et al., 2015) as reference.
The DNA methylation profile for C. rubella was obtained using previously published data (Seymour et al., 2014). Sequencing reads were remapped using Bismark (Krueger and Andrews, 2011) with -bowtie1 -n 1 option. The metagene plots (Figure 5) were constructed using DeepTools (Ramírez et al., 2014) for both sRNA and DNA methylation data. The Deeptools computematrix parameter was set to calculate the coverage mean in 50-bp windows across all genes in each group. The density plots for methylation levels in flanking regions (±1 kb) of each gene (Supplemental Figure 5) were generated by calculating the ratio of methylated Cs to the total number of Cs within 100-bp bins, filtering out bins with less than five Cs. The bin with the highest methylation value was taken as a representative value for each gene and plotted.
The 24-nucleotide sRNA sequences mapping to +2 kb upstream regions of MEGs were extracted using bedtools (Quinlan and Hall, 2010). The extracted sequences were then remapped to the C. rubella genome using BLAT with minimum score set to 10. Any position where the 24-nucleotide sRNA could be mapped was marked in the genome and connected to the original +2 kb MEG upstream position and then represented as an ideogram (Figure 6). The ideogram was created using Circos (www.circos.ca).
High-Throughput DNA and RNA Sequence and Imprinting Analysis
Adapter trimming and low-quality read removal was performed using Trimmomatic (Bolger et al., 2014). Genomic sequencing reads were aligned to the C. rubella genome v 1.0 (Phytozome) using Bowtie (Langmead et al., 2009). Reads mapping to multiple locations were discarded using -m 1 option. SNPs were identified using FreeBayes (Garrison and Marth, 2012). Default minimum coverage was set to 20 and each alternating allele should at least cover 20% of the total coverage. SNPs with more than 200 coverages were discarded.
RNA alignment was done using TopHat (Trapnell et al., 2012). The alignment was done by using -g 1 -N 2-bowtie1 option. No multiple-location mapped reads are allowed. Base counting was done by extracting the information using igvtools (Robinson et al., 2011). Imprinted genes and TEs were identified by calculating the probability of parental alleles deviating from the two maternal to one paternal ratio using χ2 testing of reads with specific parental SNPs. The minimum coverage for each gene was set at 20. Genes and TEs with a P value > 0.01 (corrected using Benjamini-Hochberg method) were discarded.
Possible maternal tissue RNA contaminations were filtered by performing a differential expression analysis between whole seeds (Rebernig et al., 2015) and endosperm samples. Simple scaling normalization was used to compare the profiles. Genes and TEs with more than 20% decreased expression level in the endosperm library compared with the whole seed sample were discarded and treated as contaminants (Supplemental Data Set 2). Validation of the imprinted genes was done using Sanger sequencing. The list of primers is available in Supplemental Table 3.
TE Identification
Repeat models for Arabidopsis and C. rubella were generated using RepeatModeler (http://www.repeatmasker.org/RepeatModeler.html). To avoid model bias toward one species, the RepeatModeler was run by joining the genomes of Arabidopsis and C. rubella in one single file. The generated repeat model was used as an input file for RepeatMasker (http://www.repeatmasker.org/) to identify TEs in C. rubella and Arabidopsis. The one-code-to-find-them-all (Bailly-Bechet et al., 2014) Perl tool was used to refine the RepeatMasker result by eliminating multiple hits, duplicated hits, and compounded hits.
To identify the potential source of enriched simple repeats in the flanking region of imprinted genes, we created a simple repeat search table (Supplemental Data Set 9) by running RepeatMasker using a model that was generated for Arabidopsis and C. rubella repeats on TE sequences from the Arabidopsis TAIR database. We tested only simple repeats that were found to be enriched (hypergeometric test, P value ≤ 0.05) within MEGs and PEGs and their flanking regions (Supplemental Data Set 6) for their association with certain TE families. We counted how many times a certain repeat and the associated TE family was called (Supplemental Data Set 7). Significance of enrichment of certain TE families with imprinted genes was tested using a hypergeometric test by calculating how frequently a certain repeat type associated with a certain TE family in flanking or coding regions of MEGs/PEGs compared with all genes.
BLAST Analysis
Potential homologs of C. rubella imprinted genes in Arabidopsis were identified using the dc-megablast algorithm (minimum e-value 1e-5; windows size 40). The most similar homolog was defined as the highest scoring gene. Meanwhile, for interspecies comparison, BLASTn algorithm was used with e-value 1e-6.
GO Analysis
GO enrichment was analyzed using AtCOEcis (Vandepoele et al., 2009). Sequence IDs for most similar Arabidopsis homologs of C. rubella imprinted genes (20 MEGs and 18 PEGs) were used as queries. P values were corrected using Benjamini-Hochberg method.
Distribution of Fitness Effects and Rate of Adaptive Evolution
Population genomic data from a range-wide sample of 12 individuals of Capsella grandiflora (Steige et al., 2015) were used to assess selection. Focusing on C. grandiflora rather than C. rubella allows more powerful analyses of selection due to its higher level of polymorphism (Slotte et al., 2010; Williamson et al., 2014).
The data set was filtered retaining only biallelic SNPs with a minimum read depth of 15×. Repeat regions and heterozygous sites were removed. Folded site frequency spectra were obtained for 4-fold synonymous and 0-fold synonymous SNPs, using only those SNPs that had less than 20% missing data, using a custom R script. Divergence data were based on a full genome alignment of Arabidopsis, Arabidopsis lyrata, and C. rubella (Steige et al., 2015). Only sites showing no difference between the two Arabidopsis species but differing from Capsella were kept. A Jukes-Cantor correction was applied for multiple hits.
DFE-alpha analyses were done using custom Perl scripts. Separate analyses were performed for all gene sets. A set of 500 randomly chosen endosperm expressed genes with sufficient polymorphisms for imprinting analysis served as nonimprinted control. Confidence intervals and P values were obtained based on analyses of 200 bootstrap data sets, with bootstrapping done per gene. Each DFE run was repeated five times with different starting values. Replicates with the highest likelihood were kept. To assess whether variation in gene density might have an effect on our DFE-alpha inference (Slotte, 2014), we selected genes among the control set to match the distribution of gene density in 50-kb windows of ASE genes. We then reanalyzed the resampled control gene set in DFE-alpha.
Accession Numbers
Accession numbers of imprinted genes identified in this study can be found in Supplemental Data Set 1. Sequence data from this article can be found in the Gene Expression Omnibus/EMBL databases under accession numbers GSE76888, GSE67359, and PRJEB12064. Genomic and transcriptomic sequencing reads for both C. rubella parental lines and reciprocal crosses are deposited as fastq files in the Gene Expression Omnibus (GSE76888). C. rubella transcriptome data from whole seeds used to filter for maternal tissue contamination was taken from Gene Expression Omnibus with accession number GSE67359. Sequencing data for sRNAs are available at the European Bioinformatics Institute under accession number PRJEB12064.
Supplemental Data
Supplemental Figure 1. Analysis of hybrid seed development.
Supplemental Figure 2. Confirmation of 10 selected MEGs and PEGs by Sanger sequencing of RT‐PCR products.
Supplemental Figure 3. Principal component analysis scatterplot for different categories of imprinted and nonimprinted genes in C. rubella.
Supplemental Figure 4. Cross-comparison of imprinted genes in five different species.
Supplemental Figure 5. Frequency distribution of methylated regions within flanking regions of imprinted genes compared with control genes.
Supplemental Figure 6. Likelihood of genes being located within 500 bp proximity of MEGs, PEGs, or all genes of C. rubella and Arabidopsis.
Supplemental Table 1. Properties of sequencing libraries used in the experiment.
Supplemental Table 2. Enriched GO categories of conserved imprinted genes in C. rubella and Arabidopsis.
Supplemental Table 3. Primer sequences used for validation of imprinted genes.
Supplemental Data Set 1. List of C. rubella imprinted genes.
Supplemental Data Set 2. False discovery rate of genes expressed in seed coat and embryo.
Supplemental Data Set 3. List of accession-biased and unstable imprinted genes in two biological replicates.
Supplemental Data Set 4. Allelic ratio and transcript level of different groups of genes.
Supplemental Data Set 5. Most similar homologs of C. rubella genes in Arabidopsis.
Supplemental Data Set 6. Association of enriched simple repeats in the vicinity of imprinted genes to different TE families.
Supplemental Data Set 7. Number of simple repeats associated with defined TEs at different positions of imprinted genes.
Supplemental Data Set 8. List of parental-specific TEs.
Supplemental Data Set 9. Association of simple repeats and TEs.
Supplementary Material
Acknowledgments
We thank Dan Halligan for providing custom Perl scripts for DFE-alpha analyses. Sequencing was performed by the SNP&SEQ Technology Platform, Science for Life Laboratory at Uppsala University, a national infrastructure supported by the Swedish Research Council (VRRFI) and the Knut and Alice Wallenberg Foundation. This research was supported by a European Research Council Starting Independent Researcher grant and a grant from Swedish Research Council (to C.K.). T.S. acknowledges funding from the Swedish Research Council, the Magnus Bergvall foundation, the Nilsson-Ehle Foundation, and the Royal Swedish Academy of Sciences. Population genetic computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under Project b2012190.
AUTHOR CONTRIBUTIONS
C.K. and M.R.H. designed the research and wrote the manuscript. M.R.H. and K.A.S. performed the research. M.R.H., C.K., B.L., and T.S. analyzed the data. C.K. and T.S. provided all the reagents and the tools for the analysis. All authors commented on the manuscript and contributed to the writing.
Glossary
- TE
transposable element
- Mya
million years ago
- SNP
small nucleotide polymorphism
- DAP
days after pollination
- MEG
maternally expressed gene
- PEG
paternally expressed gene
- GO
Gene Ontology
- DFE
distribution of negative fitness effect
References
- Aichinger E., Villar C.B.R., Farrona S., Reyes J.C., Hennig L., Köhler C. (2009). CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet. 5: e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly-Bechet M., Haudry A., Lerat E. (2014). “One code to find them all”: a Perl tool to conveniently parse RepeatMasker output files. Mob. DNA 5: 13. [Google Scholar]
- Bechsgaard J.S., Castric V., Charlesworth D., Vekemans X., Schierup M.H. (2006). The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol. Biol. Evol. 23: 1741–1750. [DOI] [PubMed] [Google Scholar]
- Belmonte M.F., et al. (2013). Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc. Natl. Acad. Sci. USA 110: E435–E444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K., Yant L., Laitinen R.A., Kim S.-T., Hollister J.D., Warthmann N., Fitz J., Weigel D. (2010). Local-scale patterns of genetic variability, outcrossing, and spatial structure in natural stands of Arabidopsis thaliana. PLoS Genet. 6: e1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond D.M., Baulcombe D.C. (2014). Small RNAs and heritable epigenetic variation in plants. Trends Cell Biol. 24: 100–107. [DOI] [PubMed] [Google Scholar]
- Calarco J.P., Borges F., Donoghue M.T.A., Van Ex F., Jullien P.E., Lopes T., Gardner R., Berger F., Feijó J.A., Becker J.D., Martienssen R.A. (2012). Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151: 194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen R.H., Pelizzola M., Schmitz R.J., Lister R., Dowen J.M., Nery J.R., Dixon J.E., Ecker J.R. (2012). Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. USA 109: E2183–E2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A., Keightley P.D. (2009). Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol. Biol. Evol. 26: 2097–2108. [DOI] [PubMed] [Google Scholar]
- Figueiredo D.D., Batista R.A., Roszak P.J., Köhler C. (2015). Auxin production couples endosperm development to fertilization. Nat Plants 1: 15184. [DOI] [PubMed] [Google Scholar]
- Foxe J.P., Slotte T., Stahl E.A., Neuffer B., Hurka H., Wright S.I. (2009). Recent speciation associated with the evolution of selfing in Capsella Proc. Natl. Acad. Sci. USA 106: 5241–5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E., Marth G. (2012). Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907. [Google Scholar]
- Gaut B.S., Doebley J.F. (1997). DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 94: 6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M., Bubb K.L., Henikoff S. (2009). Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324: 1447–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M., Missirian V., Henikoff S. (2011). Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS One 6: e23687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossmann T.I., Song B.-H., Windsor A.J., Mitchell-Olds T., Dixon C.J., Kapralov M.V., Filatov D.A., Eyre-Walker A. (2010). Genome wide analyses reveal little evidence for adaptive evolution in many plant species. Mol. Biol. Evol. 27: 1822–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.-L., Bechsgaard J.S., Slotte T., Neuffer B., Lascoux M., Weigel D., Schierup M.H. (2009). Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc. Natl. Acad. Sci. USA 106: 5246–5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T.-F., Ibarra C.A., Silva P., Zemach A., Eshed-Williams L., Fischer R.L., Zilberman D. (2009). Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T.-F., Shin J., Uzawa R., Silva P., Cohen S., Bauer M.J., Hashimoto M., Kirkbride R.C., Harada J.J., Zilberman D., Fischer R.L. (2011). Regulation of imprinted gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. USA 108: 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra C.A., et al. (2012). Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337: 1360–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Köhler C. (2012). Evolution, function, and regulation of genomic imprinting in plant seed development. J. Exp. Bot. 63: 4713–4722. [DOI] [PubMed] [Google Scholar]
- Koch M.A., Kiefer M. (2005). Genome evolution among cruciferous plants: a lecture from the comparison of the genetic maps of three diploid species--Capsella rubella, Arabidopsis lyrata subsp. petraea, and . Am. J. Bot. 92: 761–767. [DOI] [PubMed] [Google Scholar]
- Krueger F., Andrews S.R. (2011). Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J., Ma J., Swigonová Z., Ramakrishna W., Linton E., Llaca V., Tanyolac B., Park Y.-J., Jeong O.-Y., Bennetzen J.L., Messing J. (2004). Gene loss and movement in the maize genome. Genome Res. 14: 1924–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Jacobsen S.E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat E. (2010). Identifying repeats and transposable elements in sequenced genomes: how to find your way through the dense forest of programs. Heredity (Edinb) 104: 520–533. [DOI] [PubMed] [Google Scholar]
- Li J., Berger F. (2012). Endosperm: food for humankind and fodder for scientific discoveries. New Phytol. 195: 290–305. [DOI] [PubMed] [Google Scholar]
- Luo M., Taylor J.M., Spriggs A., Zhang H., Wu X., Russell S., Singh M., Koltunow A. (2011). A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet. 7: e1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maumus F., Quesneville H. (2014). Ancestral repeats have shaped epigenome and genome composition for millions of years in Arabidopsis thaliana. Nat. Commun. 5: 4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T. (2001). Arabidopsis thaliana and its wild relatives: a model system for ecology and evolution. Trends Ecol. Evol. 16: 693–700. [Google Scholar]
- Moore T., Haig D. (1991). Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 7: 45–49. [DOI] [PubMed] [Google Scholar]
- Murphy S.K., Jirtle R.L. (2003). Imprinting evolution and the price of silence. BioEssays 25: 577–588. [DOI] [PubMed] [Google Scholar]
- Pask A.J., Papenfuss A.T., Ager E.I., McColl K.A., Speed T.P., Renfree M.B. (2009). Analysis of the platypus genome suggests a transposon origin for mammalian imprinting. Genome Biol. 10: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A.H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556. [DOI] [PubMed] [Google Scholar]
- Pignatta D., Erdmann R.M., Scheer E., Picard C.L., Bell G.W., Gehring M. (2014). Natural epigenetic polymorphisms lead to intraspecific variation in Arabidopsis gene imprinting. eLife 3: e03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires N.D., Grossniklaus U. (2014). Different yet similar: evolution of imprinting in flowering plants and mammals. F1000Prime Rep. 6: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prickett A.R., Oakey R.J. (2012). A survey of tissue-specific genomic imprinting in mammals. Mol. Genet. Genomics 287: 621–630. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F., Dündar F., Diehl S., Grüning B.A., Manke T. (2014). Deeptools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42: W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebernig C.A., Lafon-Placette C., Hatorangan M.R., Slotte T., Köhler C. (2015). Non-reciprocal interspecies hybridization barriers in the Capsella genus are established in the endosperm. PLoS Genet. 11: e1005295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. (2011). Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszak P., Köhler C. (2011). Polycomb group proteins are required to couple seed coat initiation to fertilization. Proc. Natl. Acad. Sci. USA 108: 20826–20831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez D.H., Paszkowski J. (2014). Heat-induced release of epigenetic silencing reveals the concealed role of an imprinted plant gene. PLoS Genet. 10: e1004806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour D.K., Koenig D., Hagmann J., Becker C., Weigel D. (2014). Evolution of DNA methylation patterns in the Brassicaceae is driven by differences in genome organization. PLoS Genet. 10: e1004785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She W., Baroux C. (2014). Chromatin dynamics during plant sexual reproduction. Front. Plant Sci. 5: 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T. (2014). The impact of linked selection on plant genomic variation. Brief. Funct. Genomics 13: 268–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T., Foxe J.P., Hazzouri K.M., Wright S.I. (2010). Genome-wide evidence for efficient positive and purifying selection in Capsella grandiflora, a plant species with a large effective population size. Mol. Biol. Evol. 27: 1813–1821. [DOI] [PubMed] [Google Scholar]
- Slotte T., et al. (2013). The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 45: 831–835. [DOI] [PubMed] [Google Scholar]
- Steige K.A., Reimegård J., Koenig D., Scofield D.G., Slotte T. (2015). Cis-regulatory changes associated with a recent mating system shift and floral adaptation in Capsella. Mol. Biol. Evol. 32: 2501–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Onge K.R., Källman T., Slotte T., Lascoux M., Palmé A.E. (2011). Contrasting demographic history and population structure in Capsella rubella and Capsella grandiflora, two closely related species with different mating systems. Mol. Ecol. 20: 3306–3320. [DOI] [PubMed] [Google Scholar]
- Suzuki S., et al. (2007). Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 3: e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S., Renfree M.B., Pask A.J., Shaw G., Kobayashi S., Kohda T., Kaneko-Ishino T., Ishino F. (2005). Genomic imprinting of IGF2, p57(KIP2) and PEG1/MEST in a marsupial, the tammar wallaby. Mech. Dev. 122: 213–222. [DOI] [PubMed] [Google Scholar]
- Swigonová Z., Lai J., Ma J., Ramakrishna W., Llaca V., Bennetzen J.L., Messing J. (2004). Close split of sorghum and maize genome progenitors. Genome Res. 14: 1916–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon M.I., Hollister J.D., Gaut B.S. (2010). A triptych of the evolution of plant transposable elements. Trends Plant Sci. 15: 471–478. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K., Quimbaya M., Casneuf T., De Veylder L., Van de Peer Y. (2009). Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol. 150: 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters A.J., Bilinski P., Eichten S.R., Vaughn M.W., Ross-Ibarra J., Gehring M., Springer N.M. (2013). Comprehensive analysis of imprinted genes in maize reveals allelic variation for imprinting and limited conservation with other species. Proc. Natl. Acad. Sci. USA 110: 19639–19644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson R.J., Josephs E.B., Platts A.E., Hazzouri K.M., Haudry A., Blanchette M., Wright S.I. (2014). Evidence for widespread positive and negative selection in coding and conserved noncoding regions of Capsella grandiflora. PLoS Genet. 10: e1004622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff P., Weinhofer I., Seguin J., Roszak P., Beisel C., Donoghue M.T.A., Spillane C., Nordborg M., Rehmsmeier M., Köhler C. (2011). High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet. 7: e1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf J.B., Hager R. (2006). A maternal-offspring coadaptation theory for the evolution of genomic imprinting. PLoS Biol. 4: e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H.R., Dittmer T.A., Richards E.J. (2008). Three SRA-domain methylcytosine-binding proteins cooperate to maintain global CpG methylation and epigenetic silencing in Arabidopsis. PLoS Genet. 4: e1000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Dai M., Li F., Liu A. (2014). Genomic imprinting, methylation and parent-of-origin effects in reciprocal hybrid endosperm of castor bean. Nucleic Acids Res. 42: 6987–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A., Lepère G., Jay F., Wang J., Bapaume L., Wang Y., Abraham A.-L., Penterman J., Fischer R.L., Voinnet O., Navarro L. (2013). Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc. Natl. Acad. Sci. USA 110: 2389–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Li N., He W., Zhang H., Yang W., Liu B. (2016). Genome-wide screen of genes imprinted in sorghum endosperm and the roles of allelic differential cytosine methylation. Plant J. 85: 424–436. [DOI] [PubMed] [Google Scholar]
- Zheng C., Chen E., Albert V.A., Lyons E., Sankoff D. (2013). Ancient eudicot hexaploidy meets ancestral eurosid gene order. BMC Genomics 14: S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D., Gehring M., Tran R.K., Ballinger T., Henikoff S. (2007). Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39: 61–69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.