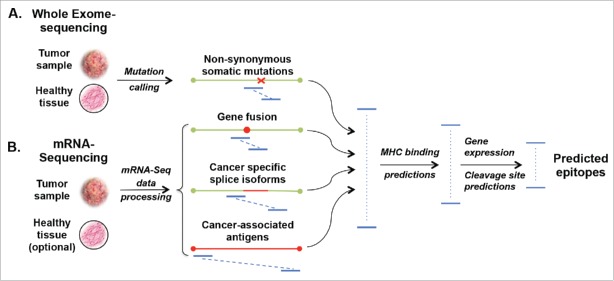
Figure 1.
Computational approaches to identify cancer-specific MHC ligands. Comparing exome sequencing data (A) from tumor and healthy tissues allows identification of non-synonymous mutations. From mRNA sequencing (B) cancer-specific gene fusions, aberrant RNA specific, and expressed/overexpressed TA genes can be identified by comparison with databases of splice isoforms and healthy tissues. All possible peptides (blue lines) containing at least one cancer-specific amino acid mutation are then subjected to MHC-binding peptide predictions. For MHC I molecules 8- to 11-mer and for MHC II 14- to 20-mer peptides are considered for predictions. Predicted peptides are ranked based on their predicted binding affinity and possibly other variables, like immunogenicity, pMHC complex stability, predicted cleavage sites or level of expression of the corresponding genes.