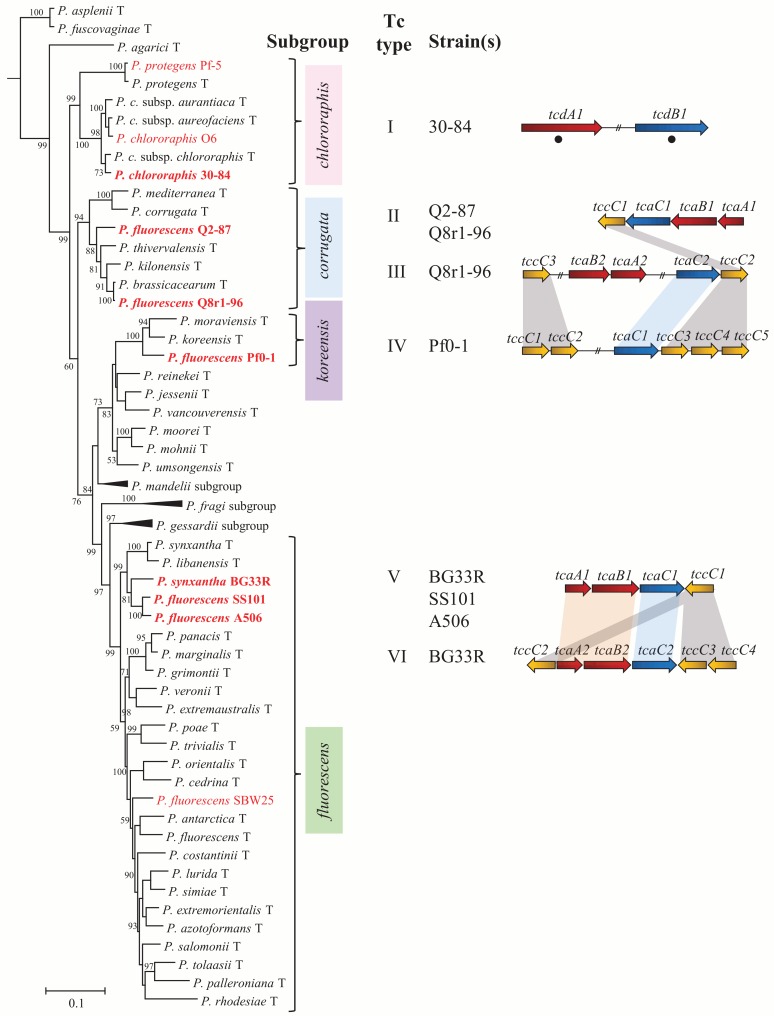
Fig 2. Seven of the ten strains of Pseudomonas spp. evaluated in this study have Tc clusters.
The ten strains evaluated in this study fall into four subgroups within the P. fluorescens group, as shown in a phylogenetic tree based on concatenated alignments of gyrB, rpoB, rpoD and 16S rRNA of the type strains within the P. fluorescens group. The ten strains examined in this study are shown in red font and subgroups containing these strains are labelled to the right (pink, chlororaphis subgroup; blue, corrugata subgroup; purple, koreensis subgroup; green, fluorescens subgroup). The tree is artificially rooted on the type strain of P. aeruginosa. Subgroups lacking any of the ten strains are collapsed and labeled. Bootstrap support less than 50% is not shown. Branch lengths indicate the number of nucleotide substitutions per site. Strains evaluated in this study that contain a Tc cluster are shown in bold, red font and are also listed to the right of the tree. Tc cluster Types I-VI are distinguished from one another by gene organization and genome location. Genes are colored according to the Tc component encoded: red (component A), blue (component B) and yellow (component C). Type I: 30–84 (Pchl3084_2947 and Pchl3084_2950); Type II: Q2-87 (PflQ2_0667–0670) and Q8r1-96 (PflQ8_0736–0739); Type III: Q8r1-96 (PflQ8_4696, PflQ8_4570–4571 and PflQ8_4580–4581); Type IV: Pf0-1 (Pfl01_0947–0948 and Pfl01_4453–4456); Type V: A506 (PflA506_3065–3068), SS101 (PflSS101_2971–2974) and BG33R (PseBG33_3189–3192); Type VI: BG33R (PseBG33_3799–3804). Numbers following gene names distinguish genes within a single genome that encode the same Tc component. Black circles denote genes located on genomic islands. Among genomes, homologous genes, defined by genomic location and phylogenetic relationships, are connected with shading of the same color.