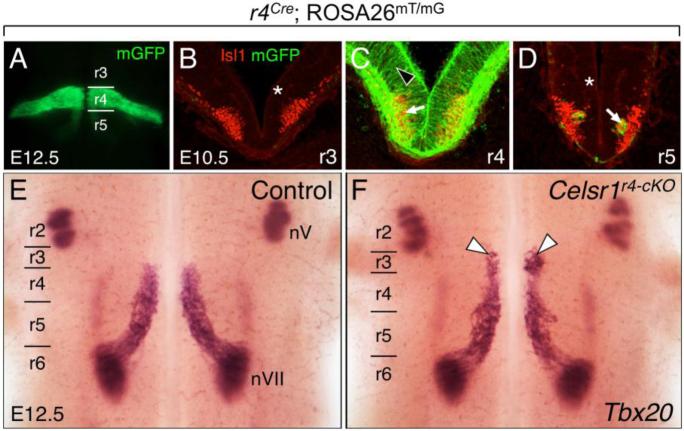
Figure 3. Some FBM neurons migrate rostrally into rhombomere 3 (r3) following disruption of Celsr1 function in r4.
A, E, F, Dorsal views of E12.5 embryos visualized live for GFP (A) or processed for Tbx20 ISH (E, F). GFP expression in r4 in a r4-Cre; ROSA26mT/mG embryo (A) labels the expression domain of Cre used for the r4-specific knockout of Celsr1 function (r4-cKO). BD, 30-μm coronal sections of an E10.5 wild type hindbrain processed for anti-GFP (green) and anti-Isl1 (red) immunostaining to mark r4-Cre expression (GFP), and FBM neurons (arrow), respectively. The light red background signal (asterisks) is due to mTomato expression in all tissues of the ROSA26mT/mG mouse where Cre is not active. Cre activity is restricted to tissues within r4 (black arrowhead). The GFP-expressing cells in D (arrow) are the earliest caudally-migrating FBM neurons originating in r4. E, F, In control embryos (r4-Cre; Celsr1fl/+ or r4-Cre; Celsr1KO/+, 20/21 embryos), Tbx20 labels trigeminal BM neurons (nV) in r2 and r3, and FBM neurons (nVII) migrating caudally from r4 to r6 (E). A small but significant number of FBM neurons (arrowheads) migrates rostrally into r3 in a majority of r4-cKO embryos (F) (r4-Cre; Celsr1KO/fl or r4-Cre; Celsr1fl/fl, 9/12 embryos).