FIGURE 2:
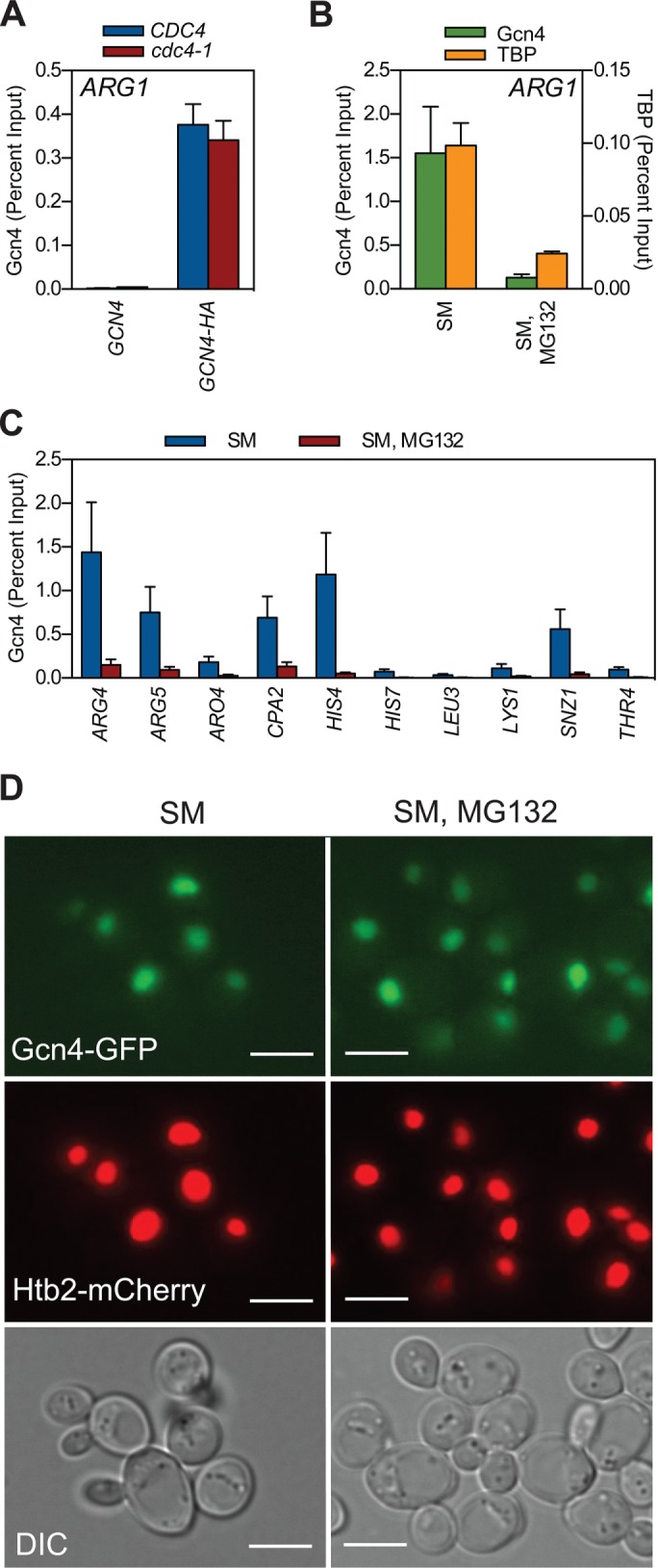
Inhibiting the proteasome impedes the ability of Gcn4 to bind target gene chromatin. (A) CDC4 (W303-1a), CDC4 GCN4-HA (GHY107), cdc4-1 (MT668), and cdc4-1 GCN4-HA (GHY107) strains were grown to log phase at 30°C in minimal medium, shifted to the restrictive temperature of 37°C for 1 h, and then induced with SM for an additional 1.5 h. At this time, ChIP was performed with an antibody against the HA-epitope tag. Coprecipitating ARG1 promoter DNA was quantified by qPCR, expressed relative to the percentage of input DNA. n = 3. (B) GCN4-HA (GHY025) yeast were grown to log phase at 30°C in minimal medium, treated with either DMSO or MG132 for 1 h, and induced with SM for 1.5 h. ChIP was performed using antibodies against the HA-epitope tag or TBP. Coprecipitating ARG1 promoter DNA was quantified by qPCR. n = 3. (C) As in B, except that coprecipitating DNAs from the anti-HA ChIP were quantified by qPCR, using primer pairs that amplify Gcn4-binding sites in the indicated genes. n = 3. Error bars represent SEM. (D) GCN4-GFP HTB2-mCherry (GHY339) yeast were grown to log phase at 30°C in minimal medium, treated with either DMSO or MG132 for 1 h, and induced with SM for 1.5 h. Samples were imaged using either fluorescence (top) or differential interference contrast microscopy (bottom). Scale bars, 5 μm.
