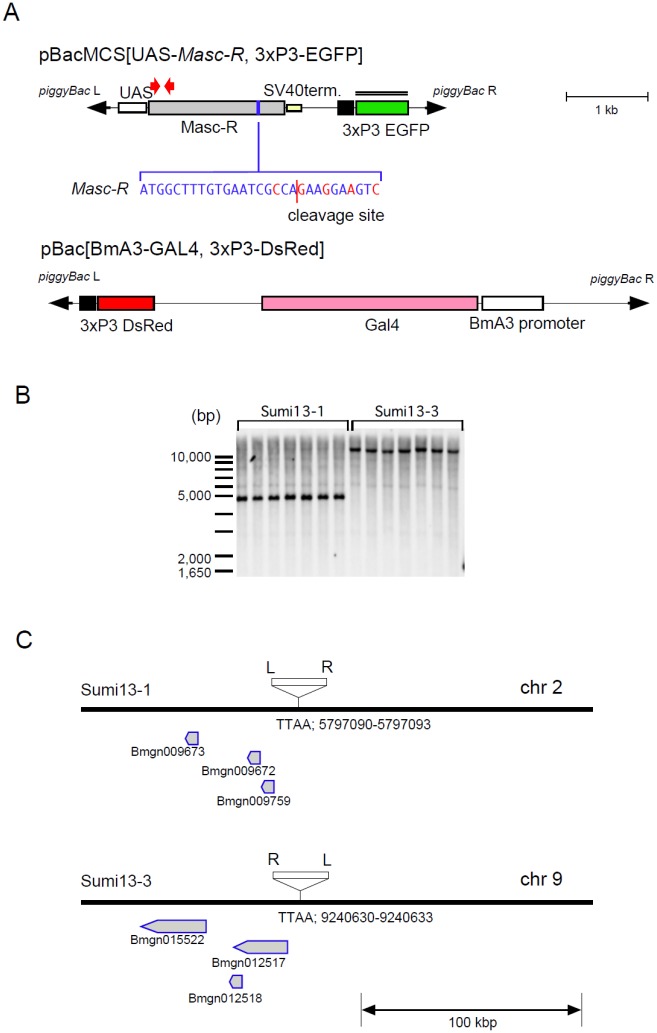
Fig 1. Generation of transgenic silkworms that express Masc-R.
(A) Construction of the piggyBac transformation vector, pBacMCS [UAS-Masc-R, 3×P3-EGFP] (in this study) and pBac [BmA3-GAL4, 3×P3-DsRed][27]. Grey box represents the open reading frame of Masc-R. Target region by Fem piRNA is shown by the blue box, and the sequences are displayed. Masc-R contains 5 nucleotide substitutions (red characters) from the wild-type sequence, which makes it resistant to cleavage by the Fem piRNA-PIWI protein complex. The red line represents the cleavage site. piggyBac inverted terminal repeats are shown as block arrows. UAS, upstream activation sequence; SV40 term, 3' untranslated region of Simian virus 40 including polyadenylation signal for the termination. The double line indicates the position of the probe used in the Southern blot analysis. The red arrows indicate the approximate location of the primers used for RT-PCR analysis described in Fig 2A. (B) Southern blot analysis of two transgenic strains (Sumi13-1 and 13–3). Genomic DNA prepared from transgenic lines was digested with BamHI and analyzed with a probe containing the EGFP sequence. The molecular size is shown on the left side of the membranes. (C) Schematic diagram of the insertion site of the transgene in each transgenic strain (Sumi13-1 and 13–3) revealed by inverse-PCR. Neighboring genes that exist around the insertion site are displayed. White box indicates the transformation vector. piggyBacL and R arms are shown as L and R, respectively. The position number of the TTAA insertion sequence within the chromosome is described below the line.