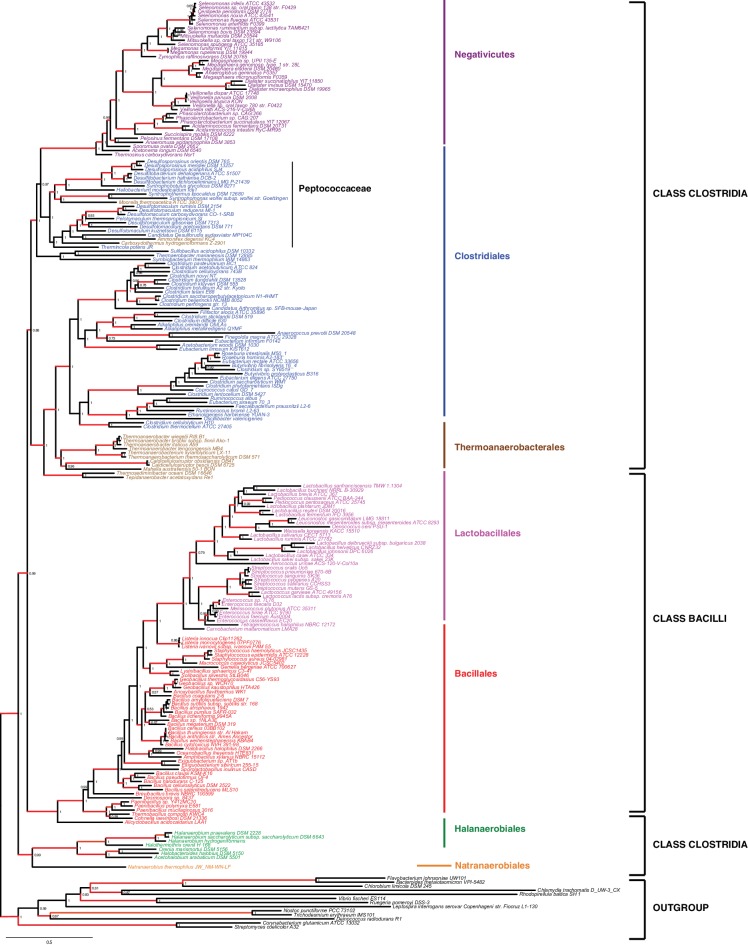
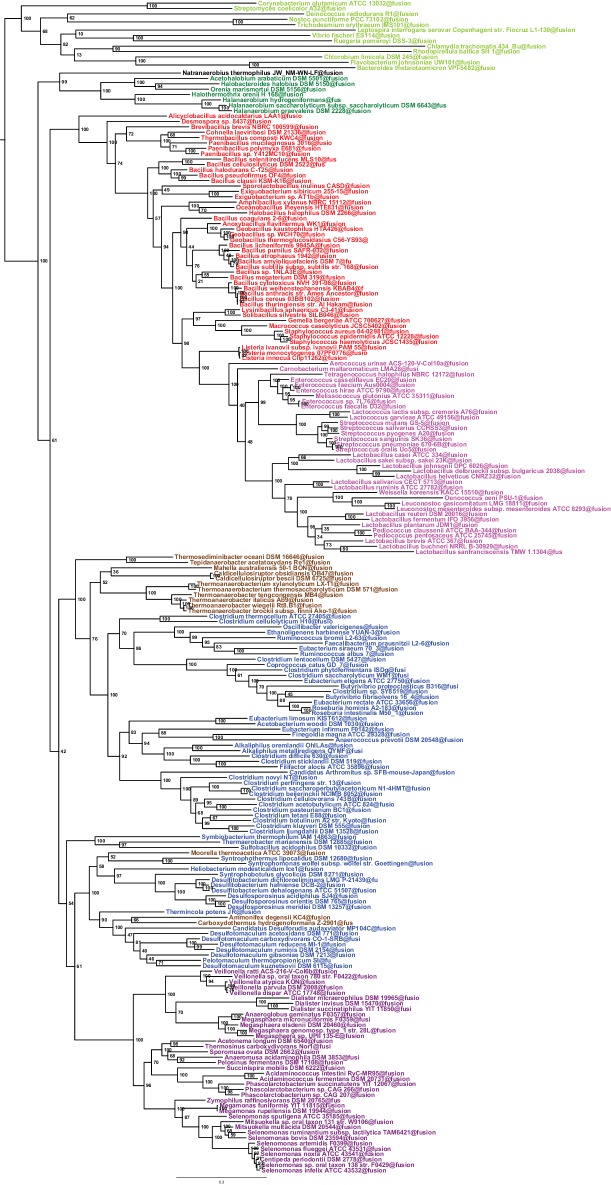
Figure 2. Phylum-level phylogeny of the Firmicutes.
Bayesian phylogeny of the Firmicutes based on a concatenation of 47 orthologous ribosomal proteins comprising 5551 amino acid positions and the CAT+GTR+Γ4 model. Values at nodes represent Bayesian posterior probabilities. The scale bar represents the average number of substitutions per site. For details on analyses, see Materials and methods.