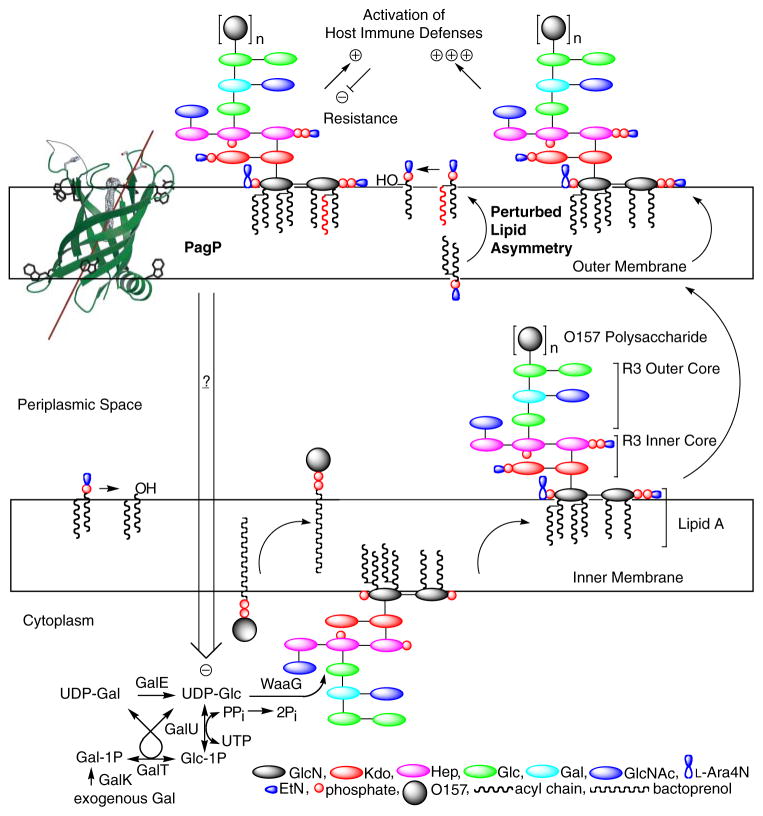
FIGURE 9. Model for PagP-mediated control of R3 core oligosaccharide structure.
PagP is the only known OM enzyme of LPS biosynthesis in E. coli, and it serves to transfer a palmitate chain from a phospholipid to lipid A with the production of a lysophospholipid by-product. Lipid A palmitoylation attenuates endotoxin signaling through the host TLR4 pathway and affords resistance to cationic antimicrobial peptides. PagP measures the phospholipid palmitate chain in its hydrocarbon ruler, which is only accessible from the OM external leaflet. A defect in cytoplasmic lipid A myristoylation triggers PagP activity in the OM of E. coli O157:H7. Activated PagP not only palmitoylates lipid A at the cell surface, to partially compensate for lipid A underacylation, but also separately initiates signal transduction through its periplasmic amphipathic α-helix. Signal transduction exerts negative control on the cytoplasmic UDP-Glc pool and leads to a truncation of the R3 core oligosaccharide.