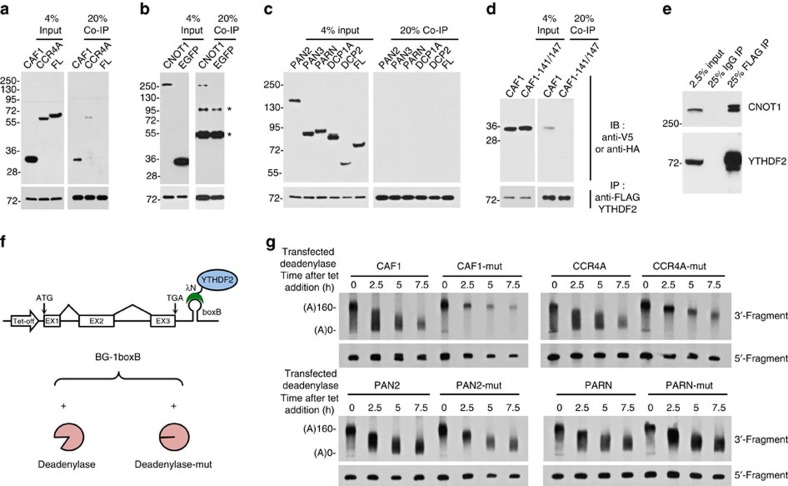
Figure 3. The CCR4–NOT complex is responsible for YTHDF2-mediated RNA deadenylation.
(a–c) Interaction between YTHDF2 and deadenylases or decapping enzymes. HEK 293 cells were co-transfected with a plasmid encoding FLAG-tagged YTHDF2 and either a V5-tagged CAF1, CCR4A, PAN2, PAN3, PARN, DCP1A, DCP2, FL or an HA-tagged CNOT1 or enhanced green fluorescent protein (EGFP). Lysates were subjected to immunoprecipitation by using anti-FLAG affinity gel. Input and co-purified proteins were blotted by probing with corresponding antibodies. Nonspecific bands were indicated by the asterisk. (d) Interaction between YTHDF2 and CAF1 or CAF1-141/147, a mutant version of CAF1 with M141K and L147K double substitution. (e) Interaction between stably expressed FLAG-tagged YTHDF2 and endogenous CNOT1 in HeLa-tTA cells. (f) Strategy of screening of functional deadenylases that are responsible for YTHDF2-mediated deadenylation. BG-1boxB reporter was co-transfected with λN-FLAG-YTHDF2 and either a wild-type or catalytically inactive deadenylase. (g) Deadenylation assay of BG-1boxB tethered by YTHDF2 with either a co-transfected wild-type or catalytically inactive deadenylase.