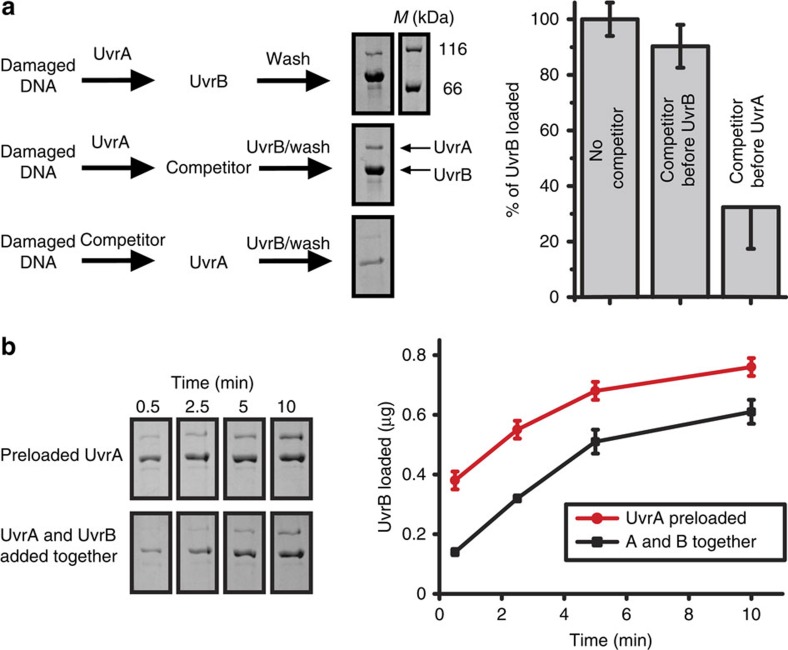
Figure 5. In vitro UvrB-loading assays.
(a) Competition assay. UvrA was incubated with biotinylated DNA, containing a site-specific fluorescein lesion, attached to magnetic beads. Subsequently, UvrB was added and the amount of UvrB recruited to damaged DNA beads was quantified. As a competitor, a 10-fold excess of damaged DNA containing the same site-specific lesion was added to the reaction, either before addition of UvrA or before addition of UvrB. Images of representative gels are presented and mean of three independent experiments is shown on the right. The total amount of UvrB protein loaded onto the beads was quantified and compared between the three conditions used. The most efficient loading reaction was set as 100%. Errors represent s.d. of three experimental repeats. For full gel images see Supplementary Fig. 4. (b) Time course experiment, which monitored the efficiency of UvrB recruitment to damaged DNA over time. Two reactions were performed with either UvrA preloaded onto the damaged DNA followed by UvrB at t=0, or premixed UvrA and UvrB were added together at t=0. Images of representative gels are presented and the mean of three independent experiments is shown on the right. Errors represent s.e.m. of three experimental repeats. For full gel images see Supplementary Fig. 4.