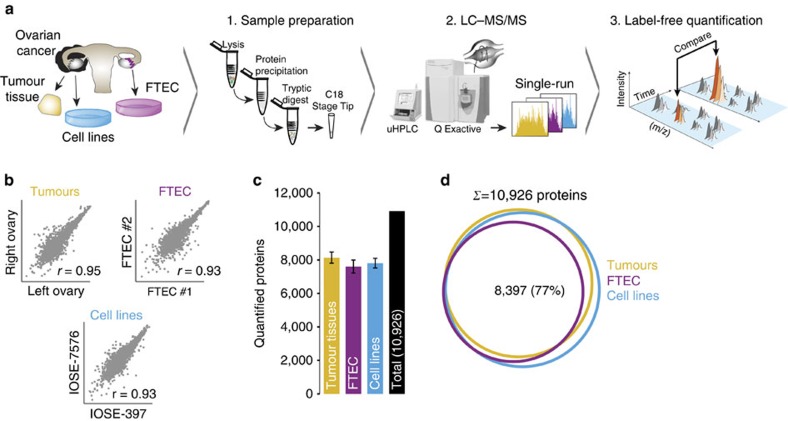
Figure 1. Deep single-run proteomics of cell lines and human ovarian cancer tissue.
(a) Summary of the shotgun proteomics workflow for OvCa cellular models and high-grade serous ovarian cancer (HGSOC) tissues. Following lysis, protein purification, and tryptic digest, peptides were separated by ultra-high performance liquid chromatography and measured in single runs using a quadrupole Orbitrap mass spectrometer. Label-free proteome quantification was performed using the MaxQuant software environment. (b) Workflow reproducibility for cell lines (n=30), HGSOC tissues (n=8) and primary FTEC cells (n=3). Pearson correlations (r) were calculated for biological replicates of cell lines, primary FTEC isolates from different healthy donors, and ovarian tumour tissues from both ovaries from a woman with HGSOC. (c) Average number of quantified proteins from each sample type. Error bars represent standard deviations. (d) Number of proteins common to FTECs, HGSOC tumours and cell lines. FTEC, fallopian tube epithelial cell; LC-MS/MS, liquid chromatography tandem mass spectrometry; m/z, mass-to-charge ratio; uHPLC, ultra-high performance liquid chromatography.