Figure 1. FATE1 is associated with mitochondria in human ACC cells.
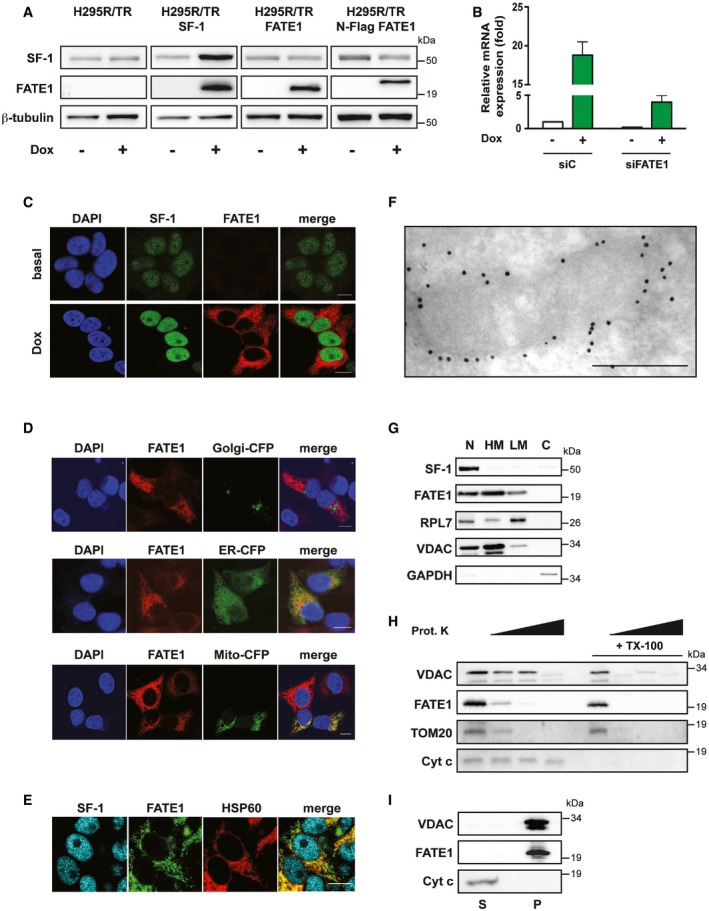
- SF‐1, FATE1 and β‐tubulin protein levels shown in basal condition and after Dox treatment in H295R/TR, H295R/TR SF‐1, H295R/TR FATE1, and H295R/TR N‐Flag FATE1 cells.
- FATE1 mRNA expression is increased in H295R/TR SF‐1 cells by Dox treatment. The efficiency of FATE1 knockdown by specific (siFATE1) vs. control (siC) siRNA nucleofection is shown (mean ± SEM; n = 4 with 3 replicates/experiment).
- Immunofluorescence showing endogenous FATE1 protein (red) induction of expression by Dox treatment of H295R/TR SF‐1 cells. SF‐1 (green), DAPI (blue). Scale bars, 10 μm.
- Subcellular localization of endogenous FATE1 (red) and transfected fluorescent markers for Golgi, ER, and mitochondria, respectively (green) in Dox‐treated H295R/TR SF‐1 cells. DAPI (blue). Scale bars, 10 μm.
- Subcellular localization of the endogenous FATE1 protein (green) in Dox‐treated H295R/TR SF‐1 cells costained with an antibody against the mitochondrial marker HSP60 (red). SF‐1 (blue). Scale bar, 10 μm.
- Immunogold electron microscopy showing association of FATE1 with the mitochondrial outer surface in Dox‐treated H295R/TR N‐Flag FATE1 cells. Scale bar, 500 nm.
- Dox‐treated H295R/TR N‐Flag FATE1 cells were fractioned into nuclear (N), heavy membranes (HM), light membranes (LM), and cytosolic (C) fractions, and localization of SF‐1, FATE1, ribosomal protein RPL7, VDAC1, and GAPDH was revealed by immunoblot.
- Effect of increasing concentrations (0, 1, 10 and 100 μg/ml) of proteinase K and 0.1% Triton X‐100 (TX‐100) treatment of the mitochondrial fraction from Dox‐treated H295R/TR N‐Flag FATE1 cells on VDAC1, FATE1, TOM20, and cytochrome c.
- FATE1 and VDAC1 are associated with the pellet (membrane; P) fraction after high‐speed centrifugation of the alkaline‐extracted mitochondrial fraction of Dox‐treated H295R/TR SF‐1 cells, while cytochrome c is found in the supernatant (S).
Source data are available online for this figure.
