Fig. 4.
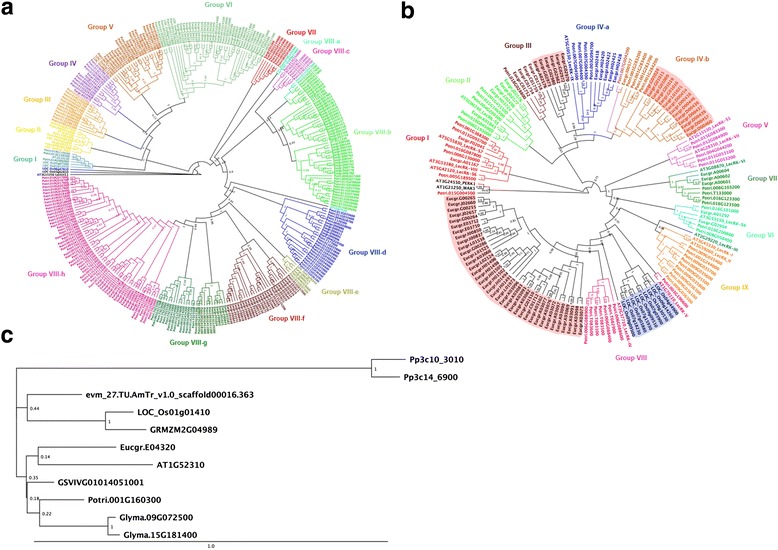
Maximum likelihood phylogenetic analysis of G-, L-, and C-type PtLecRLKs with LecRLKs from Arabidopsis, rice and Eucalyptus. The full-length amino acid sequences of G- and L-type LecRLKs were collected from Arabidopsis, rice, and Eucalyptus genome to perform phylogeny analysis with those of Populus. C-type PtLecRLK was tested with full length amino acid sequences of those of moss (Physcomitrella patens), shrub (Amborella trichopoda), corn (Zea mays), soybean (Glycine max) and grape (Vitis vinifera) as well as those of Arabidopsis, rice and Eucalyptus. a Maximum likelihood for constructing G-type LecRLKs phylogenetic tree calculated by LG + G model. aLRT branch support is displayed in each node. AT3G15356 (Lectin 3.1) was used as a distal protein to build phylogenetic tree. b Maximum likelihood phylogenetic tree with JTT + G + invariant sites (I) model of L-type LecRLKs. The distantly related AT1G21250 (WAK1) and AT3G24550 (PERK1) were rooted to classify groups in this analysis. aLRT branch support value is displayed in each node. The clades including only G-type EgLecRLKs are highlighted with light red. A clade including G-type rice LecRLK is highlighted with light blue. c Maximum likelihood phylogenetic tree of C-type LecRLKs with JTT + G model using 1000 bootstrap. Bootstrap values are shown to each node. The bar indicates the number of amino acids substitution per site
