Fig. 6.
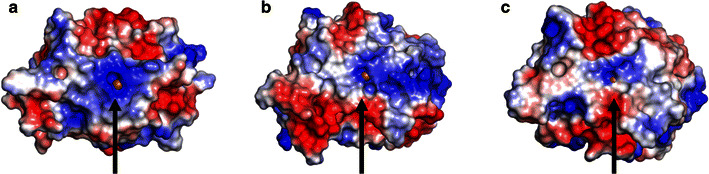
Cartoons of the surface charge distribution of the structural models of a MtLPMO9A, b MtLPMO9B and c MtLPMO9C. Protein orientation: the flat substrate-binding site (Fig. 3) is located to the front of all three LPMOs and the copper ion is indicated by the black arrow. Recent NMR studies revealed a direct interaction of the reductant CDH with a narrow surface patch in the vicinity of the copper ion [26]. MtLPMO9A is strongly positively charged in the vicinity of the copper ion compared to MtLPMO9B and MtLPMO9C based on the surface charge distribution (pH 5.0). The scaling from the negative and positive electrostatic potential regions are −5 for blue and +5 for the red regions. The electrostatic map was obtained from APBS plugin from PyMOL
