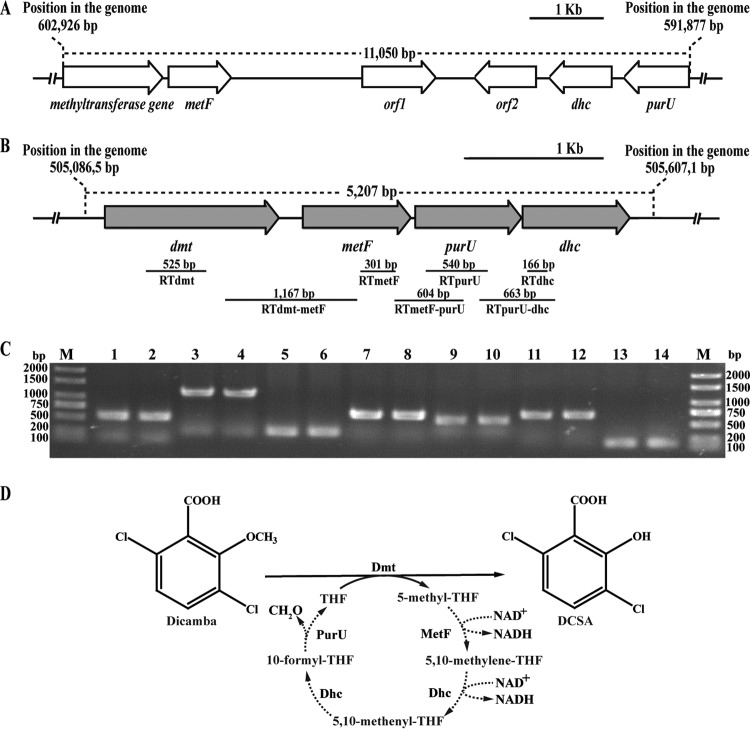
FIG 1.
(A, B) Organization of the methyltransferase gene cluster in scaffold 02 (A) and scaffold 66 (B). The gray arrows indicate the sizes and transcriptional direction of each gene in scaffold 66. The locations of the primer sets RTdmt, RTdmt-metF, RTmetF, RTmetF-purU, RTpurU, RTpurU-dhc, and RTdhc and the DNA fragments amplified by RT-PCR are indicated below. (C) Transcription analysis of dmt, metF, purU, and dhc in scaffold 66 by RT-PCR. Total RNAs of strain Ndbn-20 grown with glucose or dicamba were prepared for RT-PCR. Lane M, molecular markers; lanes 2, 4, 6, 8, 10, 12, and 14 (template from cells grown with dicamba), products obtained from the reactions of the RTdmt, RTdmt-metF, RTmetF, RTmetF-purU, RTpurU, RTpurU-dhc, and RTdhc primer sets, respectively, with the RT products. Lanes 1, 3, 5, 7, 9, 11, and 13, corresponding negative controls. (D) Proposed dicamba O-demethylation system linked to the THF-mediated C1 metabolism in strain Ndbn-20. The reactions indicated by a dotted line have not been confirmed by experiments. MetF, 5,10-methylene-THF reductase; Dhc, the bifunctional enzyme 5,10-methylene-THF dehydrogenase/5,10-methenyl-THF cyclohydrolase; PurU, formyl-THF deformylase.