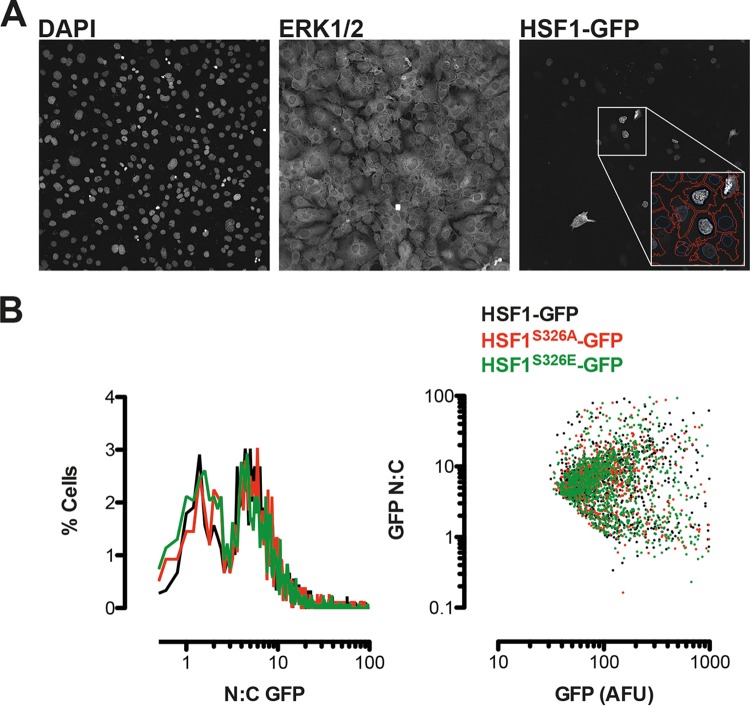
FIG 8.
S326A/E mutation does not influence the nucleocytoplasmic distribution of ectopically expressed HSF1-GFP. HSF1-knockout MEFs were transfected with GFP-tagged wild-type or S326A/E mutants of HSF1 prior to counter staining with anti-ERK1/2 antibodies and DAPI. Four fields of view per well of eight replicate wells per condition were imaged using a robotic high-content microscope. Automated and systematic analysis of images was performed using a custom algorithm. (A) A single representative field of view is shown from one well. DAPI and ERK1/2 images were used to define nuclear and cytoplasmic regions, respectively, and GFP fluorescence was recorded from each region (as indicated in the inset screengrab showing automated cell definition), accepting >70 AFU per cell as positively transfected. (B) Plots of single cell data show a frequency histogram of nuclear/cytoplasmic GFP fluorescence (left panel) indicating a bimodal distribution of HSF1, which is unaffected by S326A/E mutation. The right-hand panel shows a comparison of whole-cell GFP fluorescence in the same cell populations versus nucleocytoplasmic distribution, indicating that the bimodal distribution is apparent across a 10-fold difference in HSF1-GFP levels and is again unaffected by S326A/E substitution.