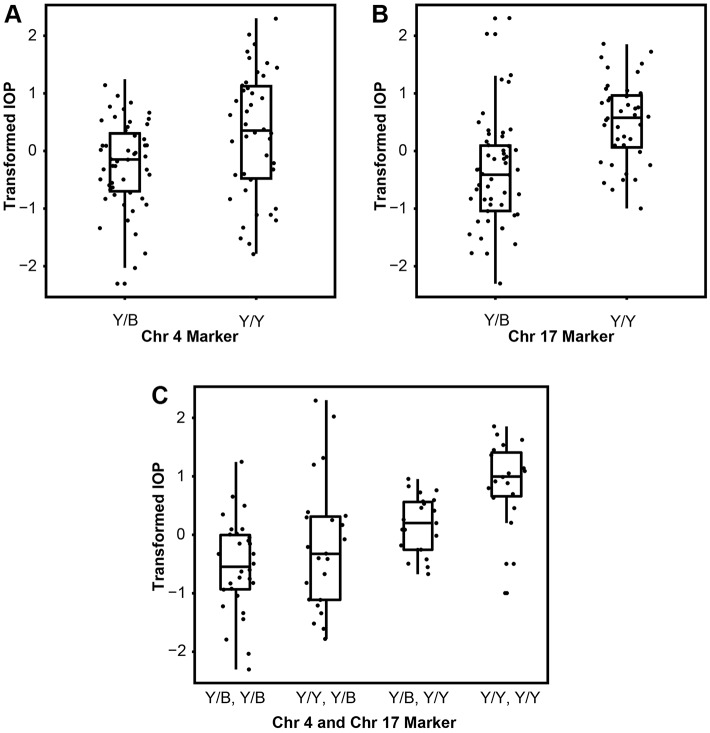
Fig. 6.
Factors independent of iris disease contribute to high IOP. Graphs showing the relationship between IOP (transformed values, see Materials and Methods) and the genotypes at the two significant QTL markers (at chromosome 4 and chromosome 17). (A,B) Mice homozygous for either a recessive YBR chromosome 4 or YBR chromosome 17 locus have significantly higher IOP than mice carrying their respective heterozygous alleles (P-value=4.239×10−3 for YBR chromosome 4 comparison and P-value=2.125×10−6 for YBR chromosome 17 comparison). (C) Mice homozygous for both the recessive YBR chromosome 4 and YBR chromosome 17 loci, respectively (Y/Y, Y/Y), have significantly higher IOP compared to mice carrying heterozygous alleles on both loci (chromosome 4 Y/B, chromosome 17 Y/B) or a recessive allele on any one of the loci (chromosome 4 Y/Y, chromosome 17 Y/B or chromosome 4 Y/B, chromosome 17 Y/Y). P=1.05×10−7 for ‘chromosome 4 Y/Y, chromosome 17 Y/Y’ vs ‘chromosome 4 Y/B, chromosome 17 Y/B’ comparison. P=5.75×10−4 for ‘chromosome 4 Y/Y, chromosome 17 Y/Y’ vs ‘chromosome 4 Y/Y, chromosome 17 Y/B’ comparison and P=1.07×10−3 for ‘chromosome 4 Y/Y, chromosome 17 Y/Y’ vs ‘chromosome 4 Y/B, chromosome 17 Y/Y’ comparison. Y and B denote YBR and B6 alleles, respectively. Chr, chromosome. Graphs are boxplots, where boxes represent data within the first and third quartiles, the whiskers represent the median values, the ends of vertical lines represent the minimum and maximum values, and the dots represent the actual data points staggered horizontally to avoid any overlaps.