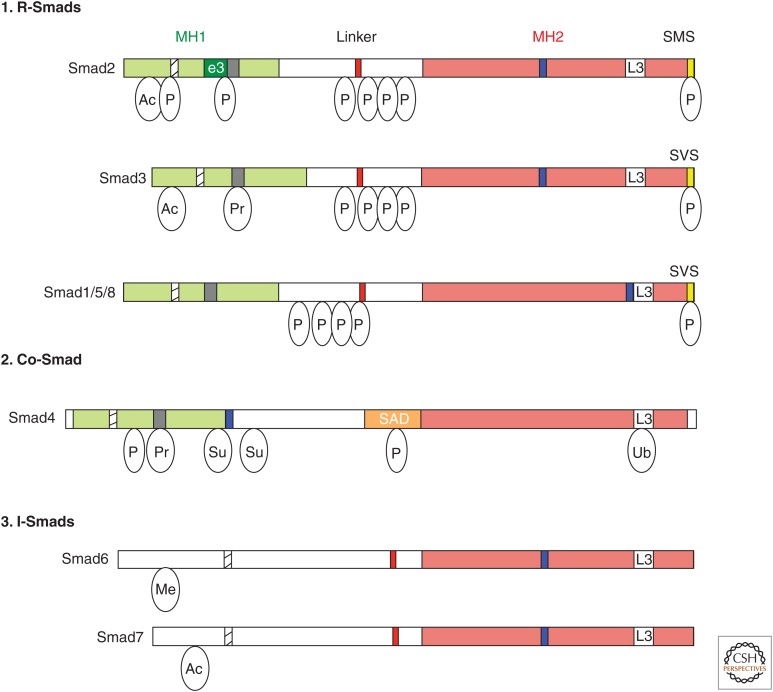
Figure 2.
The Smad family. Schematic representations of the eight human Smad proteins divided into (1) receptor-regulated Smads (R-Smads), (2) common-Smad (co-Smad), and (3) inhibitory-Smads (I-Smads). The conserved amino-terminal Mad-homology 1 (MH1) and carboxy-terminal MH2 domains are indicated as green and red boxes, respectively. Highlighted are the nuclear localization signal (NLS, hatched box), the unique insert in Smad2-MH1 domain, which corresponds to exon 3 (e3, dark green box), the β-hairpin in the MH1 domain that binds DNA or the stem region of a subset of primary microRNAs (pri-miRNAs) (black box), the proline-tyrosine (PPXY) motif (red box) in the linker domain that is recognized by the WW domain of Smurf family proteins, the Smad activation domain (SAD, orange box) at the linker-MH2 border of Smad4, the nuclear export signal (NES, blue box), and the L3 loop of the MH2 domain (white box). The carboxy-terminal serine residues in the SXS motif that is phosphorylated by the type I receptor kinases are shown in the yellow box. Relative locations of different posttranslational modifications identified in Smad proteins are indicated. Ac, Acetylation; Ub, ubiquitylation; Pr, poly(ADP)ribosylation; Su, sumoylation; P, Ser or Thr phosphorylation.