FIG 2.
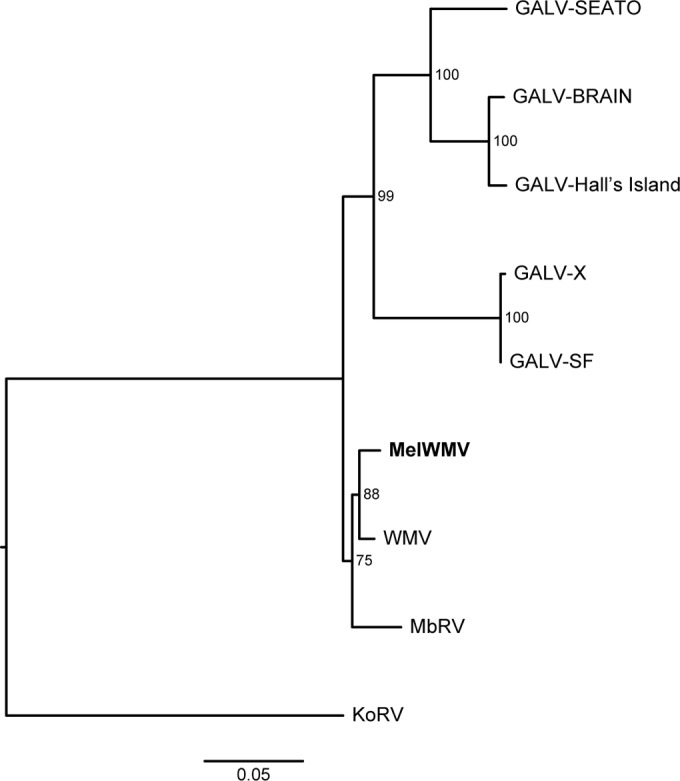
Maximum likelihood phylogenetic tree of GALVs inferred using concatenated partitioned full-genome nucleotide sequences. Coding sequences, noncoding LTRs, and intergene spacers were included in the analysis. The sequences obtained from GenBank with the corresponding accession codes are GALV-SEATO (KT724048), GALV-SF (KT724047), GALV-Brain (KT724049), GALV-Hall's Island (KT724050), woolly monkey virus (WMV; KT724051), and Melomys burtoni retrovirus (MbRV; KF572483 to KF572486). The MelWMV sequence generated in study is shown in bold. KoRV (AF151794) was used as the outgroup. Node support was assessed with 500 rapid bootstrap pseudoreplicates and is indicated at each node. The scale bar indicates 0.05 nucleotide substitutions per site. The tree is midpoint rooted for purposes of clarity.
