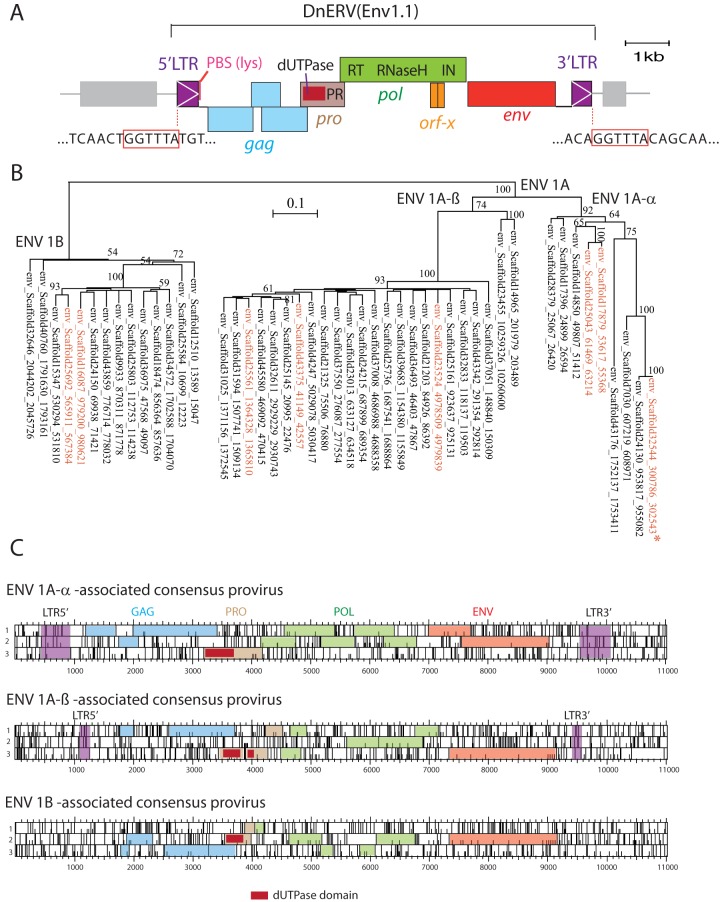
FIG 6.
Characterization of the DnERV(Env1.1) provirus and related sequences. (A) The DnERV(Env1.1) provirus, with the gag, pro, pol, and env genes; the identified typical domains (including the characteristic betaretroviral pro-N-terminal dUTPase domain); and the proviral 5′ and 3′ LTRs indicated. Repeated long interspersed nuclear elements (LINE) retrotransposons (gray), as identified by the RepeatMasker Web program, are positioned. The 6-bp target site duplication flanking the provirus (red boxes), a characteristic feature of retroviral integration, is indicated. An ∼400-bp region downstream of the env gene (corresponding to part of the 3′ LTR) was PCR amplified and sequenced to fill the unresolved gap (a series of N′s) in the Dasnov3.0 database. In addition, the 5′ and 3′ LTRs of the DnERV(Env1.1) provirus were PCR amplified and sequenced from the genomes of 5 U.S. D. novemcinctus individuals heterozygous for the proviral insertion. PBS, primer-binding site. (B) Maximum likelihood tree with the TM subunit amino acid sequences of the identified dasy-env1.1-related sequences in the armadillo genome. The vertical branch length is proportional to the percentage of amino acid substitutions from the node (bar on the top), and the percent bootstrap values (>50%) obtained from 1,000 replicates are indicated at the nodes. In red are the eight previously identified env sequences encoding full-length ORFs (Fig. 1). The asterisk indicates the functional dasy-env1.1 gene. (C) Consensus sequences generated from an alignment of the provirus sequences containing dasy-env1.1-related genes for each of the three phylogenetic groups determined as described above for panel B (ORF maps). The gag, pro, pol, and env genes with the characteristic betaretroviral pro-N-terminal dUTPase domain and the proviral 5′ and 3′ LTRs (when present) are indicated.