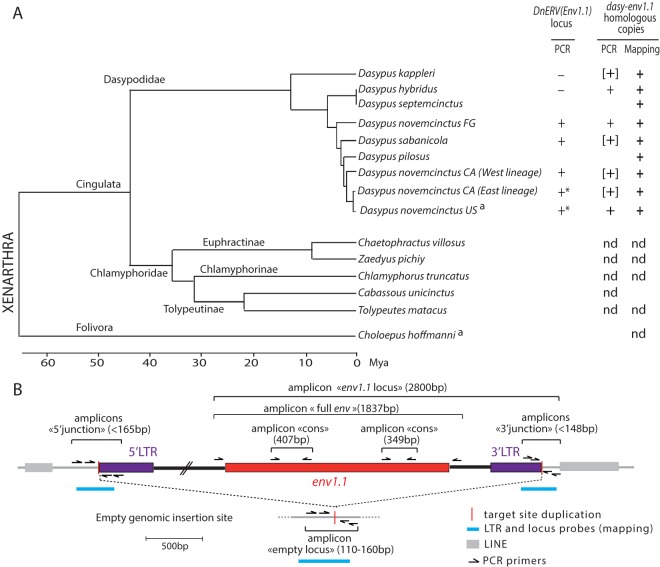
FIG 9.
Identification and conservation of dasy-env1.1 and its related elements during xenarthran radiation. (A, left) Phylogenetic tree of Xenarthra (31) with the names of the species tested together with their corresponding families and subfamilies. The horizontal branch length is proportional to time. FG, French Guiana; CA, Central America. a, species whose genome is available in genomic databases. (Right) The presence (+) or absence (−) of the DnERV(Env1.1) orthologous locus or of dasy-env1.1 homologous copies determined by using either PCR assays (“PCR”) or mapping on genomic Illumina reads (“Mapping”) is indicated for each species tested (the individual status of each species is provided in Table 1). Brackets indicate that only a partial sequence could be retrieved. Asterisks indicate that a full-length dasy-env1.1 ORF was detected at the orthologous locus in individuals of this species (Table 1). nd, not detected. (B) Strategy used to detect the DnERV(Env1.1) provirus at its orthologous locus as well as provirus-related sequences. The presence or absence of a proviral locus was determined by using three PCRs (with a limited elongation time): the “5′ junction” and “3′ junction” primer pairs amplify sequences when DnERV is present at the locus. If the provirus is not integrated at the locus, the “empty locus” primer pair, but not the two other primer combinations, yields a PCR product (corresponding to the joined flanking sequences). If all 3 primer pairs yield a product, the provirus insertion is heterozygous. If no primer pairs amplify a sequence, the quality of the DNA may be too poor. The presence of dasy-env1.1 at the orthologous locus was determined by using the “env1.1 locus” primer pair. The presence or absence of DnERV(Env1.1)-related sequences was determined by using three PCRs with two “cons” primer pairs and the “full env” primer pair, which amplify partial and full-length env fragments, respectively. The schematized amplicons are not drawn to scale. Finally, the positions of the probes that were used for the mapping of the shotgun Illumina genomic reads are indicated.