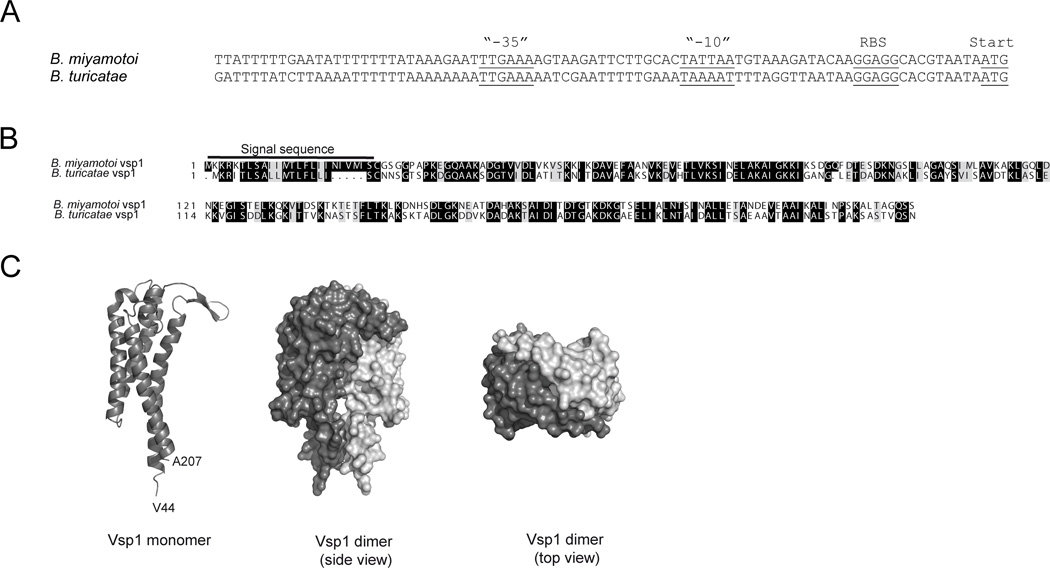
FIGURE 2. Borrelia miyamotoi Vsp1 promoter region and predicted protein structure.
(A) Alignment of the promoter regions of the B. miyamotoi and B. turicatae expression sites, based on B. miyamotoi Vsp1 (KF031441.1) and B. turicatae VspB (AF049852) sequences. AT-rich sequences surround the “-35” promoter element. RBS: ribosomal binding site. Start: start codon. (B) Alignment of B. miyamotoi Vsp1 and B. turicatae Vsp1 amino acid sequences, including their respective signal sequences, conserved amino acids (grey background) and identical residues (black background). (C) Structure prediction based on annotated crystal structures of B. turicatae Vsp1 and B. burgdorferi OspC using the Phyre2 webportal. Vsp1 and OspC are known to form dimers, and in this figure a predicted monomer (dark gray) has been aligned to a B. turicatae Vsp1 dimer crystal structure (PDB 2GA0), consisting of 2 B. miyamotoi Vsp1 monomers (light and dark grey, respectively).