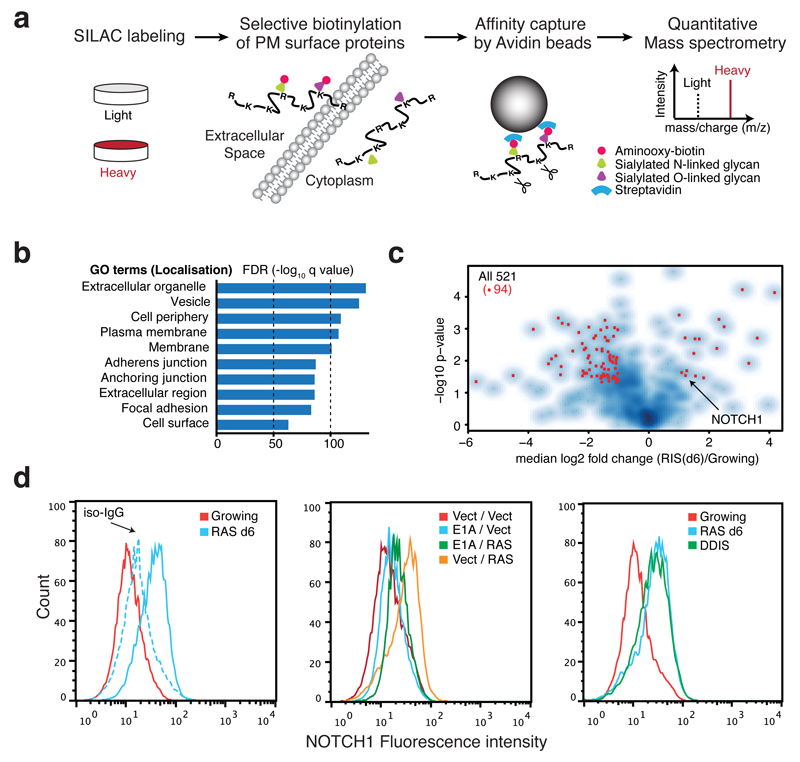
Figure 1. Plasma membrane proteomics (PMP) defines NOTCH1 as upregulated in OIS.
(a) The workflow for quantitative PMP using differential SILAC labelling of growing and HRASG12V-induced senescent (RIS) IMR90 cells. (b) GO cellular compartment term enrichment for all 1502 identified proteins in both conditions. (c) Volcano plot of 521 high-confidence protein identifications from PMP demonstrating log2 fold change (RIS(d6) / Growing) against negative log10 p value (n = 4 independent experiments). Among 167 proteins differentially expressed during RIS (p<0.05), red dots indicate 94 proteins with more than two fold change. (d) Cell surface NOTCH1 expression by flow-cytometry in indicated IMR90 cells: left, ER:HRASG12V cells with (d6) or without (Growing) 4OHT, iso-IgG, isotype control IgG; centre, cells with constitutive overexpression of either HRASG12V, E1A, or both; right, DNA damage-induced senescence (DDIS). To establish DDIS, cells were treated with 100μM Etoposide for 2 days, followed by 5-days incubation in drug-free medium.