FIGURE 5.
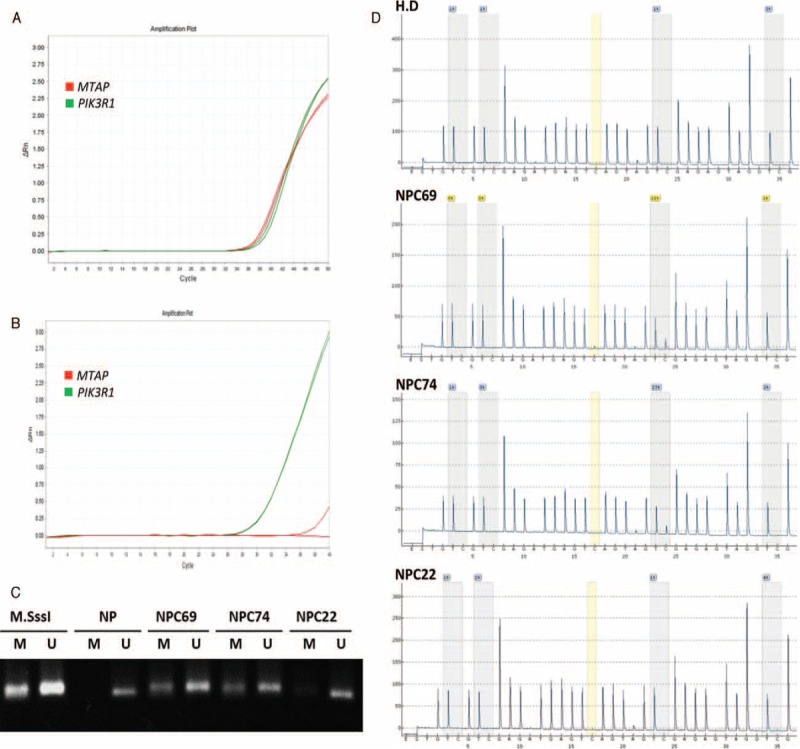
MTAP gene dosage and methylation-specific PCR for analysis of MTAP promoter methylation in nasopharyngeal carcinoma. Representative low-stage NPC with intact MTAP expression has a preserved MTAP gene, with a Ct value similar to that of PIK3R1, both <40 in a quantitative DNA-PCR (A). Representative high-stage and MTAP-deficient NPC has nondetectable MTAP gene, characterized by a Ct value no less than 40 (B). Methylation-specific PCR detects MTAP promoter methylation in MTAP protein-deficient NPCs without homozygous gene deletion (C). PCR products amplified from MTAP promoter with methylation-specific primers can be distinctly visualized in both NPC69 and NPC74, while the MTAP gene promoter of a representative MTAP-intact NPC, NPC22, is unmethylated. Pyrosequencing is performed to quantify the percentage of methylation in specific CpG sites of MTAP (D). The percentages of methylation in NPC69 and NPC74 are 23% and 26%, respectively. While in NPC22, the percentage is 4%, which is less than the cut-off value of methylation and regarded as unmethylated. M = methylation-specific primers, M.SssI = a healthy donor's blood DNA treated with M. SssI methyltransferase, MTAP = methylthioadenosine phosphorylase, NP = DNA from nontumor nasopharyngeal mucosal tissue without treatment of M.SssI methyltransferase, NPC = nasopharyngeal carcinoma, PCR = polymerase chain reaction, U = unmethylation-specific primers.
