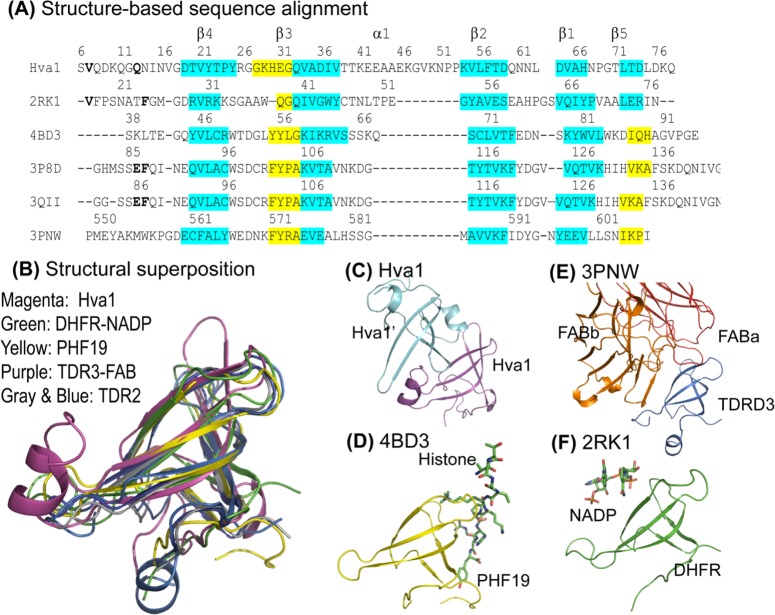
Fig 5. Structural homology analysis.
(A) This structure-based sequence comparison was generated by aligning the five PDB structures over the coordinates of Hva1. Consistent with Fig 4, the sequence corresponding to β-strands and the α-helix are labelled β1–5 and α1 respectively. The regions with high superposition (RMSD < 0.5 Å) are highlighted in cyan while the regions of the conserved β-strands with lesser overlap (0.5–2.0 Å) are distinguished in yellow. The PDB structures used in this figure are: 2RK1 –Tetrameric type II dihydrofolate reductase complexed with dihydrofolate. 4BD3 –PHF19 bound with Histone H3. 3P8D –Homodimeric Tudor 2 domain of human PHF 20. 3QII–Tudor domain 2 of human PHD finger protein 20. 3PNW–Tudor domain of human TDRD3 complexed with anti-TDRD3 FAB. (B) The structural superposition of the six structures reveals the striking tertiary homology between the proteins, especially the β-strands. The structural comparison also revealed that the same interface is used for protein-ligand and protein-protein interactions: (C) intermolecular interaction in Hva1; (D) PHF19 (yellow) bound with histone (stick model) (4BD3); (E) TDRD3 (purple) complexed with FAB (orange and red chains) (3PNW); and (F) dihydrofolate reductase (green) co-crystallized with NADP (stick model) (2RK1).