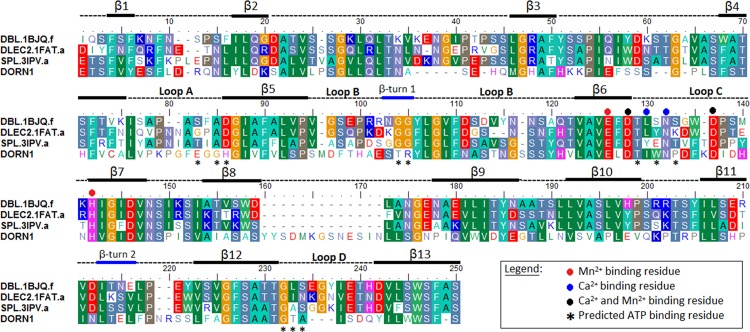
Fig 3. Multiple sequence-structural alignment of the DORN1 L-type lectin domain and three selected templates.
Template sequence names were assigned by the Uniprot database, following by a 4-letter PDB code and chain name, separated by a dot. DBL-Dolichos biflorus (1BJQ), DLEC2-Phaseolus vulgaris (1FAT), SPL-Spatholobus parviflorus (3IPV), and DORN1-Arabidopsis thaliana (P2K1). Numbers at the beginning and the end of the alignment denote actual amino acid position in the protein sequences. Black bars-beta strands, blue bars-beta turns, dash lines-loops or coils, * predicted ATP binding residue, + conserved sugar binding residue inferred from legume lectins, red dots-Mn2+ binding residues, blue dots-Ca2+ binding residues, black dots-Mn2+ and Ca2+ binding residues. Columns with same color indicate identical amino acids or similar groups of amino acids, dashes are gap insertions. The black bars (beta strands) and blue bars (beta turns) below the alignment represent the secondary structure of the DORN1 L-type lectin domain inferred from the modeled DORN1 structure. Round dots with different colors below the aligned columns are cation binding residues inferred from the templates. Black stars denoted predicted ATP binding residues of the DORN1. Loops A, B, C and D and an extended loop inferred from the alignment with legume lectins.