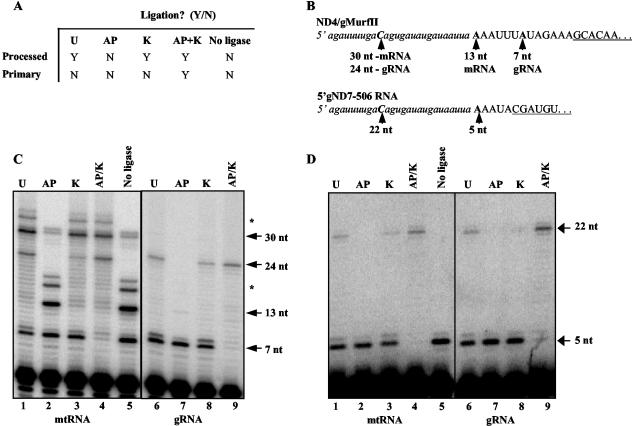
FIG. 4.
Ligation-dependent poisoned primer extension assay of total mtRNA and gel-purified gRNA. (A) Table of expected ligation outcomes for processed and primary transcripts subjected to various enzymatic treatments. Treatments: U, untreated; K, kinase; AP, alkaline phosphatase; AP/K, alkaline phosphatase followed by kinase; no ligase, untreated RNA not subjected to ligation. (B) Nucleotide sequence of the synthetic RNA oligonucleotide (lowercase italics) and the 5′ ends of ND4-gMURF2-II and gND7(506) (capital letters). The 3′ ends of the primers used in the extension reactions are underlined and the sizes of the expected products are indicated by arrows. Primer extension products were electrophoresed in 8% acrylamide-urea gels. (C) The 7-nt unligated gMURF2-II product is visible for all treatments but the positive control (AP/K), while the 13-nt unligated ND4 product is present only in the negative control lane (AP). (D) Minicircle-encoded gND7(506) remains unligated for all but the positive control lane (AP/K). Asterisks in panel C indicate the minor mRNA 5′ ends seen in Fig. 2A and the corresponding ligation products.